Serum proteins and faecal microbiota as potential biomarkers in newly diagnosed, treatment-naïve inflammatory bowel disease and irritable bowel syndrome patients
- PMID: 40481885
- PMCID: PMC12343743
- DOI: 10.1007/s00109-025-02558-5
Serum proteins and faecal microbiota as potential biomarkers in newly diagnosed, treatment-naïve inflammatory bowel disease and irritable bowel syndrome patients
Abstract
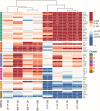
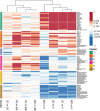
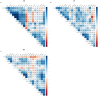
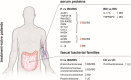
Molecular biomarkers are valuable tools to predict the disease and determine its course. Several markers have been associated with inflammatory bowel disease (IBD) and irritable bowel syndrome (IBS); however, none is sufficiently reliable to enable accurate diagnosis. We characterized a broad panel of serum proteins to assess disease-specific biomarker profiles and associate these findings with faecal microbiota composition in newly diagnosed IBD and IBS patients and healthy individuals. The study cohort consisted of 49 newly diagnosed treatment-naïve adult patients (13 Crohn's disease (CD), 13 ulcerative colitis (UC), and 23 IBS) and 12 healthy individuals. Inflammatory and metabolism-related serum proteins were assessed using PEA multiplex panels, while gut microbiota composition was determined by 16 s rRNA gene amplicon sequencing. Serum proteins AXIN1, TNFSF14, RNASE3, EN-RAGE, OSM, ST1A1, CA13 and NADK were identified as markers with the most promising specificity/sensitivity and predictivity between healthy and disease groups, while IL-17A and TNFRSF9 enabled differentiation between IBD and IBS patients. Increased abundance of Enterobacteriaceae was associated with protein markers significantly elevated in IBD/IBS. In contrast, depletion of beneficial taxa like Ruminococcaceae and Verucomicrobiaceae (i.e. Akkermansia muciniphila) was associated with decrease of a set of markers in diseased groups. Differences in the abundance of Turicibacteriaceae were more predictive to discern CD from UC than any of the serum proteins investigated. By using a broad panel of inflammation and metabolism-related proteins, we determined serum markers with significantly different levels in treatment-naïve IBD and IBS patients compared to healthy individuals, as well as between IBD and IBS. KEY MESSAGES : Significant changes in the levels of several serum proteins and abundances of faecal bacterial taxa between study groups were found. Increased levels of AXIN1, TNFSF14, RNASE3, EN-RAGE, OSM, ST1A1, CA13 and NADK characterize both IBD and IBS, while IL-17A and TNFRSF9 differentiate IBD from IBS. Increase of Enterobacteriaceae and depletion of beneficial taxa Ruminococcaceae and Verucomicrobiaceae (i.e. Akkermansia muciniphila) was found in IBD and IBS. Differences in Turicibacteriaceae were more predictive to discern CD from UC than any of the serum proteins investigated.
Keywords: Crohn’s disease; Faecal microbiota; Inflammatory bowel disease; Irritable bowel syndrome; Serum biomarkers; Treatment-naïve patients; Ulcerative colitis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval and consent to participate: The study was approved by the competent institutional ethics committees (380–59-10106–14-55/149, 641–01/14–02/01; 02/21/JG, 8.1.-14/45–2) and conducted in accordance with the Declaration of Helsinki. Informed consent was obtained from all individual participants included in the study. Competing interests: The authors declare no competing interests.
Figures

References
-
- Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL et al (2018) Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 390:2769–2778. 10.1016/s0140-6736(17)32448-0 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

