Identification of therapeutic targets in lung adenocarcinoma using Mendelian randomization and multi-omics
- PMID: 40481979
- PMCID: PMC12145347
- DOI: 10.1007/s12672-025-02835-2
Identification of therapeutic targets in lung adenocarcinoma using Mendelian randomization and multi-omics
Abstract
Background: Lung adenocarcinoma (LUAD) remains associated with limited effective pharmacological treatment options. This study aimed to identify potential therapeutic targets for LUAD through the integration and analysis of multi-omics datasets.
Methods: A meta-analysis was conducted using two extensive proteomics datasets, the UK Biobank Proteomics Project (UKB-PPP) and the Fenland study, to identify disease-associated targets for LUAD through the Summary-Data-Based Mendelian Randomization method. Sensitivity analysis, including heterogeneity tests for dependent instruments, were conducted to validate the findings. The prognostic relevance of the identified candidate targets was assessed using transcriptomic data. Functional interactions were explored via protein-protein interaction network analysis, while single-cell analyses were employed to determine cell-specific expression patterns and differentiation trajectories. Potential side effects and therapeutic indications of these targets were evaluated using phenome-wide association studies and pharmacological data mining.
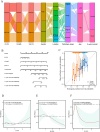
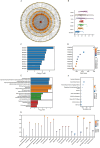
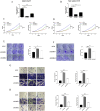
Results: Following meta-analysis, a primary significant target, intercellular adhesion molecule 5 (ICAM5), along with potential targets FUT8 and KLK13, were identified as therapeutic candidates for LUAD. FUT8 demonstrated a positive association with LUAD risk (OR = 1.02, p = 0.049), while ICAM5 (OR = 0.88, p = 0.002) and KLK13 (OR = 0.85, p = 0.021) exhibited negative associations. ICAM5 was further identified as an independent prognostic factor for patient survival (HR: 0.788, 95% CI: 0.663-0.936, p = 0.007) and revealed significant diagnostic and prognostic utility in LUAD. ICAM5 expression correlated with various immune infiltration patterns, suggesting potential modulation of the tumor immune microenvironment. Single-cell analysis revealed that ICAM5 did not directly impact LUAD cell differentiation, though its downstream target, MUC1, may contribute to differentiation processes, particularly in KRAS-mutated LUAD. Furthermore, phenome-wide association studies did not reveal substantial evidence of adverse phenotypes linked to ICAM5, supporting its safety profile for drug development.
Conclusion: ICAM5 emerges as a promising biological marker with significant prognostic and therapeutic potential in LUAD.
Keywords: Drug target; ICAM5; LUAD; Mendelian randomization; Prognosis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Systematic proteome-wide Mendelian randomization using the human plasma proteome to identify therapeutic targets for lung adenocarcinoma.J Transl Med. 2024 Apr 4;22(1):330. doi: 10.1186/s12967-024-04919-z. J Transl Med. 2024. PMID: 38576019 Free PMC article.
-
Exploring new drug treatment targets for immune related bone diseases using a multi omics joint analysis strategy.Sci Rep. 2025 Mar 27;15(1):10618. doi: 10.1038/s41598-025-94053-7. Sci Rep. 2025. PMID: 40148470 Free PMC article.
-
Involvement of ICAM5 in Carcinostasis Effects on LUAD Based on the ROS1-Related Prognostic Model.J Inflamm Res. 2024 Sep 20;17:6583-6602. doi: 10.2147/JIR.S475088. eCollection 2024. J Inflamm Res. 2024. PMID: 39318995 Free PMC article.
-
Identifying Lipid Metabolism-Related Therapeutic Targets and Diagnostic Markers for Lung Adenocarcinoma by Mendelian Randomization and Machine Learning Analysis.Thorac Cancer. 2025 Mar;16(6):e70020. doi: 10.1111/1759-7714.70020. Thorac Cancer. 2025. PMID: 40107973 Free PMC article.
-
SMR-guided molecular subtyping and machine learning model reveals novel prognostic biomarkers and therapeutic targets in non-small cell lung adenocarcinoma.Sci Rep. 2025 Jan 10;15(1):1640. doi: 10.1038/s41598-025-85471-8. Sci Rep. 2025. PMID: 39794414 Free PMC article.
References
-
- Gridelli C, Rossi A, Carbone DP, Guarize J, Karachaliou N, Mok T, Petrella F, Spaggiari L, Rosell R. Non-small-cell lung cancer. Nat Rev Dis Primers. 2015;21(1):15009. 10.1038/nrdp.2015.9. (PMID: 27188576). - PubMed
-
- Leiter A, Veluswamy RR, Wisnivesky JP. The global burden of lung cancer: current status and future trends. Nat Rev Clin Oncol. 2023;20(9):624–39. 10.1038/s41571-023-00798-3. (Epub 2023 Jul 21 PMID: 37479810). - PubMed
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. 10.3322/caac.21708. (Epub 2022 Jan 12 PMID: 35020204). - PubMed
-
- Xu H, Jiang L, Qin L, Shi P, Xu P, Liu C. Single-cell transcriptome analysis reveals intratumoral heterogeneity in lung adenocarcinoma. Environ Toxicol. 2024;39(3):1847–57. 10.1002/tox.24048. (Epub 2023 Dec 22 PMID: 38133212). - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
