An mRNA vaccine encoding the SARS-CoV-2 Omicron XBB.1.5 receptor-binding domain protects mice from the JN.1 variant
- PMID: 40482468
- PMCID: PMC12177156
- DOI: 10.1016/j.ebiom.2025.105794
An mRNA vaccine encoding the SARS-CoV-2 Omicron XBB.1.5 receptor-binding domain protects mice from the JN.1 variant
Abstract
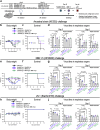
Background: The SARS-CoV-2 Omicron BA.2.86 variant and its descendant lineages, including JN.1, are rapidly spreading globally. We developed mRNA encoding the SARS-CoV-2 RBD derived from XBB.1.5 (XBB.1.5-type LNP-mRNA-RBD), in line with WHO recommendations. Many individuals have acquired immunity specific to the ancestral SARS-CoV-2 strain or early Omicron variants, such as BA.1, BA.2, or BA.5, through natural infection and/or vaccination. However, the efficacy of XBB.1.5-type LNP-mRNA-RBD boost vaccination against a clinical isolate of JN.1 remains uncertain.
Methods: In this study, we used a small amount of LNP-mRNA-RBD as a prime dose compared with a booster shot to mimic the waning immunity against the ancestral and BA.4/5 strains. We immunised female mice with XBB.1.5-type LNP-mRNA-RBD as a booster vaccine and examined the cellular and humoural responses as well as the protective efficacy against a JN.1 variant.
Findings: We found that immunisation of mice with the XBB.1.5-type LNP-mRNA-RBD as a booster shot induced XBB.1.5-specific neutralising activity and T cell responses. Moreover, immunisation with a bivalent vaccine consisting of the ancestral-type and BA.4/5-type LNP-mRNA-RBD as the primary dose followed by XBB.1.5-type LNP-mRNA-RBD boosting induced enhanced levels of cross-reactive antibodies against the JN.1 strain, compared to using the ancestral-type vaccine as the primary dose. In addition, we found that a booster shot of LNP-mRNA-RBD based on the XBB.1.5 strain reduced the viral burden in the respiratory organs after JN.1 challenge.
Interpretation: Our findings suggest that XBB.1.5-type LNP-mRNA-RBD is effective against antigenically distinct JN.1 infection.
Funding: This work was supported by grants from the Japan Program for Infectious Diseases Research and Infrastructure (JP25wm0125002), the Japan Initiative for World-leading Vaccine Research and Development Centers (JP253fa627001), and the Vaccine Development project (JP21nf0101625) from the Japan Agency for Medical Research and Development, and the National Institutes of Allergy and Infectious DiseasesCenter for Research on Influenza Pathogenesis and Transmission (CRIPT) (75N93021C00014).
Keywords: LNP-mRNA-RBD; Mouse model; Omicron JN.1 variant; SARS-CoV-2.
Copyright © 2025 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests Y. Kawaoka has received funds in the form of grants from the Japan Program for Infectious Diseases Research and Infrastructure (JP25wm0125002), the Japan Initiative for World-leading Vaccine Research and Development Centers (JP253fa627001), and the Vaccine Development project (JP21nf0101625) from the Japan Agency for Medical Research and Development, and the National Institutes of Allergy and Infectious Diseases Center for Research on Influenza Pathogenesis and Transmission (CRIPT) (75N93021C00014). N.J., K.M., and F.T. are employees of Daiichi Sankyo Co., Ltd. Y.K. is a co-founder of FluGen and has received unrelated funding support from Daiichi Sankyo Co., Ltd., Fujifilm Toyama Chemical Co., Ltd., Tauns Laboratories, Inc., Shionogi & Co. Ltd., Otsuka Pharmaceutical Co., Ltd., KM Biologics Co. Ltd., Kyoritsu Seiyaku Corporation, Shinya Corporation, and Fuji Rebio, Inc. M. Suthar serves as an advisor for Ocugen, Inc. S.Y. received royalties from Daiichi Sankyo Co., Ltd. T.J.S. L. is the founder and CEO of Nezu Biotech GmbH, Heidelberg, Germany. The remaining authors declare that they have no competing interests.
Figures


References
-
- Yang S., Yu Y., Jian F., et al. Antigenicity and infectivity characterisation of SARS-CoV-2 BA.2.86. Lancet Infect Dis. 2023;23(11):e457–e459. - PubMed
-
- Yang S., Yu Y., Xu Y., et al. Fast evolution of SARS-CoV-2 BA.2.86 to JN.1 under heavy immune pressure. Lancet Infect Dis. 2024;24(2):e70–e72. - PubMed
-
- Wang Q., Guo Y., Liu L., et al. Antigenicity and receptor affinity of SARS-CoV-2 BA.2.86 spike. Nature. 2023;624(7992):639–644. - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

