Spatial and genomic profiling of residual breast cancer after neoadjuvant chemotherapy unveil divergent fates for each breast cancer subtype
- PMID: 40482645
- PMCID: PMC12208320
- DOI: 10.1016/j.xcrm.2025.102164
Spatial and genomic profiling of residual breast cancer after neoadjuvant chemotherapy unveil divergent fates for each breast cancer subtype
Abstract
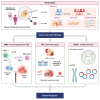
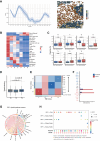
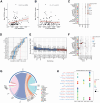
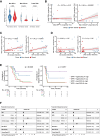
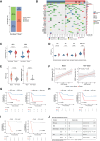
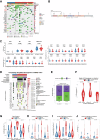
Residual cancer burden (RCB) is a strong prognostic marker after neoadjuvant chemotherapy (NAC) in breast cancer (BC), yet some BCs defy their predicted outcomes. Using single-cell spatial transcriptomics and genomic profiling, we investigate mechanisms underlying divergent fates of BCs with high RCB across subtypes. In triple-negative BC (TNBC), CXCL9+ macrophage-CD8+ T cell interactions via chemokines and interferon-gamma signaling promote favorable outcomes, while SPP1+ macrophage-cancer cell interactions driven by hypoxia signaling correlate with poor prognosis. In non-TNBC, the extent of basal-like cancer cells and their proximity to scarce immune cells are linked to prognosis. Additionally, tumor-intrinsic features-such as homologous recombination deficiency in hormone receptor (HR)-positive cancers and structural variations, including extrachromosomal ERBB2 DNA in human epidermal growth factor receptor 2 (HER2)-positive cancers-predict worse outcomes. This study highlights distinct genomic and microenvironmental strategies governing BC subtype-specific fates after NAC.
Keywords: breast cancer; extrachromosomal DNA; intrinsic subtype; neoadjuvant treatment; residual cancer burden; spatial transcriptomics; whole-genome sequencing.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests W.-Y.P. is the founder and chief executive officer of Geninus Inc., and E.S.S. is an employee of Geninus Inc.
Figures


References
-
- Loibl S., André F., Bachelot T., Barrios C.H., Bergh J., Burstein H.J., Cardoso M.J., Carey L.A., Dawood S., Del Mastro L., et al. Early breast cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2024;35:159–182. doi: 10.1016/j.annonc.2023.11.016. - DOI - PubMed
-
- Cortazar P., Zhang L., Untch M., Mehta K., Costantino J.P., Wolmark N., Bonnefoi H., Cameron D., Gianni L., Valagussa P., et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384:164–172. doi: 10.1016/S0140-6736(13)62422-8. - DOI - PubMed
-
- Symmans W.F., Wei C., Gould R., Yu X., Zhang Y., Liu M., Walls A., Bousamra A., Ramineni M., Sinn B., et al. Long-Term Prognostic Risk After Neoadjuvant Chemotherapy Associated With Residual Cancer Burden and Breast Cancer Subtype. J. Clin. Oncol. 2017;35:1049–1060. doi: 10.1200/JCO.2015.63.1010. - DOI - PMC - PubMed
-
- Kim J.Y., Oh J.M., Lee S.K., Yu J., Lee J.E., Kim S.W., Nam S.J., Park Y.H., Ahn J.S., Kim K., Im Y.H. Improved Prediction of Survival Outcomes Using Residual Cancer Burden in Combination With Ki-67 in Breast Cancer Patients Underwent Neoadjuvant Chemotherapy. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.903372. - DOI - PMC - PubMed
-
- Yau C., Osdoit M., van der Noordaa M., Shad S., Wei J., de Croze D., Hamy A.S., Laé M., Reyal F., Sonke G.S., et al. Residual cancer burden after neoadjuvant chemotherapy and long-term survival outcomes in breast cancer: a multicentre pooled analysis of 5161 patients. Lancet Oncol. 2022;23:149–160. doi: 10.1016/S1470-2045(21)00589-1. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

