Metabolic markers detect early ostedifferentiation of mesenchymal stem cells from multiple donors
- PMID: 40483499
- PMCID: PMC12145603
- DOI: 10.1186/s13287-025-04419-x
Metabolic markers detect early ostedifferentiation of mesenchymal stem cells from multiple donors
Abstract
Background: Mesenchymal stem cells (MSC) are pivotal bioengineering tools, offering significant promise for applications in bone regeneration. However, their therapeutic potential is limited by inter-donor variability and experimental issues. This study aimed to identify robust metabolic markers of osteodifferentiation applicable across multiple donors, while providing insight into the metabolic pathways actively involved in the process.
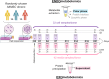
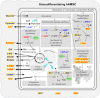
Methods: Untargeted nuclear magnetic resonance (NMR) metabolomics was applied to characterize the intra- and extracellular metabolic adaptations of human adipose-derived MSC (hAMSC) undergoing osteogenic differentiation, compared to proliferation alone. Multivariate and univariate statistical analysis was carried out on data from three independent donors, and cross-validation was employed to evaluate the predictive capacity of the proposed markers.
Results: Variations in the levels of selected (nine) intracellular and (seventeen) extracellular metabolites detect osteodifferentiation by day 7 (out of 21), with nearly 100% accuracy. These signatures suggest a metabolic shift from glycolysis/OxPhos to lactic fermentation, fatty acid β-oxidation and phosphocreatine hydrolysis. Intracellular glucose, lactate, citrate and specific amino acids are redirected towards protein synthesis and glycosylation, with some of the secreted metabolites (e.g., citrate) seemingly involved in biomineralization and other extracellular roles. Membrane metabolism, antioxidant mechanisms and adenosine metabolism are also impacted by osteodifferentiation.
Conclusions: These findings reveal effective donor-independent markers of hAMSC osteodifferentiation, with a robust extracellular signature standing out for potential rapid and non-invasive detection of osteocommitted cells.
Keywords: Bone regeneration; Mesenchymal stem cells; Metabolic markers; Metabolomics; Nuclear magnetic resonance (NMR) spectroscopy; Osteogenesis; Osteogenic differentiation.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Adipose tissue from donor 1 was obtained under an agreement between the University of Aveiro and “Hospital da Luz”, Aveiro, dated 17th February 2023 and upon written consent from the donor. The human adipose-derived stem cells (donors 2 and 3) were purchased from the American Type Culture Collection (Lot 70017032, Ref. ATCC PCS-500-011) and Lonza (Lot 22TL018258, Ref. LSLZPT-5006) having been accompanied by Certificates of Analysis stating that cells were isolated from donated human tissue after obtaining permission for their use in research applications by informed consent or legal authorization. Regarding osteoblasts, these were purchased from Lonza (CC-2538, Lot 19TL217387), certified to have been isolated from donated human tissue after obtaining permission for their use in research applications. All human cells were used within the framework of the BetterBone project (2022.04286.PTDC, doi: https://doi.org/10.54499/2022.04286.PTDC ) Human ethics and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: Authors declare that they have no competing interests. Clinical trial number: Not applicable.
Figures



References
MeSH terms
Substances
Grants and funding
- CICECO-Aveiro Institute of Materials, UIDB/50011/2020 project (doi: 10.54499/UIDB/50011/2020), UIDP/50011/2020 (doi: 10.54499/UIDP/50011/2020) and LA/P/0006/2020 (doi:10.54499/LA/P/0006/2020)/Fundação para a Ciência e a Tecnologia
- BetterBone project (2022.04286.PTDC, doi: 10.54499/2022.04286.PTDC)/Fundação para a Ciência e a Tecnologia
- Portuguese National NMR Network (RNRMN), supported by Infrastructure Project Nº 022161/Fundação para a Ciência e a Tecnologia
- SFRH/BD/150655/2020, doi: 10.54499/SFRH/BD/150655/2020/Fundação para a Ciência e a Tecnologia
- project "CellFi", PTDC/BTM-ORG/3215/2020 (DOI10.54499/PTDC/BTM-ORG/3215/2020)/Fundação para a Ciência e a Tecnologia
LinkOut - more resources
Full Text Sources

