Donor-specific digital twin for living donor liver transplant recovery
- PMID: 40486178
- PMCID: PMC12141195
- DOI: 10.1093/biomethods/bpaf037
Donor-specific digital twin for living donor liver transplant recovery
Abstract
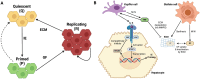
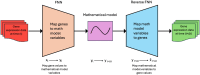
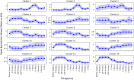
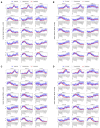
The remarkable capacity of the liver to regenerate its lost mass after resection makes living donor liver transplantation a successful treatment option. However, donor heterogeneity significantly influences recovery trajectories, highlighting the need for individualized monitoring. With the rising incidence of liver diseases, safer transplant procedures and improved donor care are urgently needed. Current clinical markers provide only limited snapshots of recovery, making it challenging to predict long-term outcomes. Following partial hepatectomy, precise liver mass recovery requires tightly regulated hepatocyte proliferation. We identified distinct gene expression patterns associated with liver regeneration by analyzing blood-derived gene expression measurements from twelve donors followed over a year. Using a deep learning-based framework, we integrated these patterns with a mathematical model of hepatocyte transitions to develop a personalized, progressive mechanistic digital twin-a virtual liver model that predicts donor-specific recovery trajectories. Central to our approach is a mechanistically identifiable latent space, defined by variables derived from a physiologically grounded differential equation model of liver regeneration, which enables biologically interpretable, bidirectional mapping between gene expression data and model dynamics. This approach integrates clinical genomics and computational modeling to enhance post-surgical care, ensuring safer transplants and improved donor recovery.
Keywords: deep learning; digital twin; liver regeneration; living donor liver transplant (LDLT); mathematical modeling; partial hepatectomy.
Published by Oxford University Press 2025.
Conflict of interest statement
The authors declare no competing interests.
Figures



Update of
-
Donor-Specific Digital Twin for Living Donor Liver Transplant Recovery.bioRxiv [Preprint]. 2025 Feb 27:2025.02.21.639518. doi: 10.1101/2025.02.21.639518. bioRxiv. 2025. Update in: Biol Methods Protoc. 2025 May 10;10(1):bpaf037. doi: 10.1093/biomethods/bpaf037. PMID: 40568069 Free PMC article. Updated. Preprint.
References
-
- Michalopoulos GK, Bhushan B. Liver regeneration: biological and pathological mechanisms and implications. Nat Rev Gastroenterol Hepatol 2021;18:40–55. - PubMed
-
- Forbes SJ, Newsome PN. Liver regeneration—mechanisms and models to clinical application. Nat Rev Gastroenterol Hepatol 2016;13:473–85. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
