NMB promotes the progression of colorectal cancer by regulating the NF-κB/P65 signaling pathway
- PMID: 40486508
- PMCID: PMC12141226
- DOI: 10.3389/fimmu.2025.1596451
NMB promotes the progression of colorectal cancer by regulating the NF-κB/P65 signaling pathway
Abstract
Background: Colorectal carcinoma (CRC) ranks as the fourth most prevalent malignancy globally. Neuromedin B (NMB) and its corresponding receptor have been implicated in the pathogenesis of multiple neoplastic conditions. Nevertheless, the specific involvement of NMB in CRC progression remains poorly characterized.
Methods: To investigate the differential expression of NMB, data were extracted from the TCGA and GEO databases, supplemented by tissue microarrays derived from clinical specimens. Prognostic significance was assessed through Kaplan-Meier survival analysis, ROC curve evaluation, Cox proportional hazards regression, clinical correlation studies, and nomogram construction. Experimental validation of NMB expression in CRC was performed using qRT-PCR, western blotting, immunohistochemistry, and ELISA techniques. Functional characterization of NMB's tumor-regulatory capacity was conducted through colony formation assays, MTT proliferation tests, wound healing experiments, and transwell migration/invasion assays. Mechanistic insights were obtained by identifying upstream regulatory proteins via co-immunoprecipitation and exploring potential signaling pathways through gene set enrichment analysis (GSEA) combined with western blot validation.
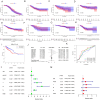
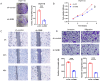
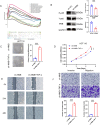
Results: Analysis of clinical data revealed that NMB exhibits markedly elevated expression levels in CRC tissues, with higher expression correlating significantly with poor prognosis. Receiver operating characteristic curve analysis validated NMB's utility as a reliable prognostic biomarker for survival outcomes. Importantly, multivariate Cox regression analysis demonstrated NMB's independent prognostic value in CRC patients. In vitro functional studies provided compelling evidence that NMB promotes CRC cell proliferation, migration, and invasive potential. Molecular investigations uncovered that NMB protein stability is regulated through USP21-dependent deubiquitination, leading to subsequent activation of the NF-κB signaling cascade and tumor progression. Notably, NMB expression patterns showed significant correlation with 5-fluorouracil resistance profiles, indicating its potential role in mediating chemotherapeutic response in CRC treatment regimens.
Conclusion: NMB exhibits significant overexpression in colorectal cancer, demonstrating promising potential as a dual-functional biomarker for both diagnostic identification and prognostic evaluation in CRC cases.
Keywords: NMB; biomarker; colorectal cancer; deubiquitination; prognosis.
Copyright © 2025 Fan, Liu, Wang, Yao, Bai, Ma, Chen, Chen, Gao, Guo and Huang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Figures






Similar articles
-
ITGB5 is a prognostic factor in colorectal cancer and promotes cancer progression and metastasis through the Wnt signaling pathway.Sci Rep. 2025 Mar 18;15(1):9225. doi: 10.1038/s41598-025-93081-7. Sci Rep. 2025. PMID: 40097546 Free PMC article.
-
High Expression Level of TRIP6 is Correlated with Poor Prognosis in Colorectal Cancer and Promotes Tumor Cell Proliferation and Migration.Biochem Genet. 2025 Feb;63(1):261-280. doi: 10.1007/s10528-024-10711-x. Epub 2024 Mar 2. Biochem Genet. 2025. PMID: 38430448
-
PFAS promotes colorectal cancer progression via regulating RIG-I-mediated innate immune signalling.Mol Immunol. 2024 Dec;176:73-83. doi: 10.1016/j.molimm.2024.11.002. Epub 2024 Nov 24. Mol Immunol. 2024. PMID: 39586166
-
UBR1 is a prognostic biomarker and therapeutic target associated with immune cell infiltration in gastric cancer.Aging (Albany NY). 2024 Aug 23;16(16):12029-12049. doi: 10.18632/aging.206079. Epub 2024 Aug 23. Aging (Albany NY). 2024. PMID: 39181686 Free PMC article.
-
Pin2/TRF1‑binding protein X1 inhibits colorectal cancer cell migration and invasion in vitro and metastasis in vivo via the nuclear factor‑κB signaling pathway.Oncol Rep. 2018 Sep;40(3):1533-1544. doi: 10.3892/or.2018.6570. Epub 2018 Jul 13. Oncol Rep. 2018. Retraction in: Oncol Rep. 2023 Feb;49(2):40. doi: 10.3892/or.2023.8477. PMID: 30015978 Retracted.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

