Global phylogenetic analysis and emergence of carbapenem-resistant Escherichia fergusonii carrying bla NDM-5 in clinic
- PMID: 40486757
- PMCID: PMC12143760
- DOI: 10.1016/j.onehlt.2025.101069
Global phylogenetic analysis and emergence of carbapenem-resistant Escherichia fergusonii carrying bla NDM-5 in clinic
Abstract
Background: Escherichia fergusonii, a newly recognized Enterobacterale member, has gained attention due to the bla NDM-5 resistance gene found in animals. However, to date, no clinical isolates carrying this gene have been reported.
Methods: A carbapenem-resistant clinical isolate of E. fergusonii was obtained from a tertiary hospital in Zhejiang Province, China. Antimicrobial susceptibility testing, filter mating, whole-genome sequencing (WGS), plasmid stability, and fitness analysis were performed. Global phylogenetic relationships of E. fergusonii were analyzed using publicly available databases.
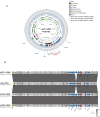
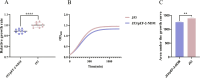
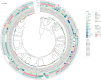
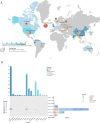
Results: E. fergusonii isolate EF21023765 exhibited resistance to most antibiotics, except tigecycline, colistin, and amikacin. WGS revealed bla NDM-5 located on an IncX3 plasmid pEF-2-NDM with a genetic environment (Tn3-IS3000-IS5-bla NDM-5-ble MBL-trpF-dsbD-IS26-ISKox3), closely related to the previously reported plasmid pEC463-NDM5, a carbapenem resistance plasmid identified from a blood sample of Escherichia coli collected in 2016 at the same Chinese hospital. This finding demonstrates the 5-year persistence of related IncX3 plasmids across distinct Escherichia species, specifically from E. coli to E. fergusonii. The plasmid pEF-2-NDM demonstrated a relatively stable state in the plasmid stability experiment. Growth rate analysis revealed a significant fitness cost associated with the bla NDM-5-carrying IncX3 plasmid, as evidenced by a 6 % higher growth rate in E. coli J53 compared to the transconjugant strain J53/pEF-2-NDM (1.00 ± 0.02 vs 0.94 ± 0.02). Phylogenetic analysis revealed significant genetic diversity in E. fergusonii, predominantly found in the UK, China, and the US, with livestock and avian as the primary hosts. In addition, ST12029 was the most prevalent sequence type.
Conclusion: To our knowledge, this is the first report of an IncX3 type plasmid carrying bla NDM-5 from a clinical E. fergusonii isolate. Our findings not only reveal the cross-species transmission capability of E. fergusonii, but also underscore its potential as an emerging pathogen, highlighting the need for enhanced surveillance to monitor the spread of bla NDM-5-producing isolates.
Keywords: Escherichia fergusonii; IncX3; Multi-drug resistant; NDM-5.
© 2025 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
References
-
- Ba X., Guo Y., Moran R.A., Doughty E.L., Liu B., Yao L., Li J., He N., Shen S., Li Y., van Schaik W., McNally A., Holmes M.A., Zhuo C. Global emergence of a hypervirulent carbapenem-resistant Escherichia coli ST410 clone. Nat. Commun. 2024;15:494. doi: 10.1038/s41467-023-43854-3. - DOI - PMC - PubMed
-
- Reference Gene Catalog - Pathogen Detection - NCBI 2025. https://www.ncbi.nlm.nih.gov/pathogens/refgene/#NDM (accessed December 24, 2024)
LinkOut - more resources
Full Text Sources
Miscellaneous

