Deciphering cargo contents in extracellular vesicles of Candida haemulonii var. vulnera
- PMID: 40487199
- PMCID: PMC12141846
- DOI: 10.1016/j.csbj.2025.04.014
Deciphering cargo contents in extracellular vesicles of Candida haemulonii var. vulnera
Abstract
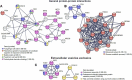
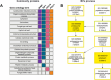
Candida haemulonii comprises a group of pathogenic fungi with notable resistance to antifungal treatments and diagnostic challenges. The Candida haemulonii variety vulnera is of particular clinical importance due to its multidrug resistance and association with invasive infections, particularly in immunocompromised patients such as diabetics and neonates. Moreover, it has been implicated in hospital outbreaks, posing a significant challenge for infection control and antifungal treatment. Extracellular vesicles (EVs) released by these fungi play critical roles in pathogen-host interactions, potentially influencing antifungal resistance, immune evasion, and virulence. Previous studies conducted by our group have demonstrated that EVs carry immunogenic cargo that can influence the host's immune response. A comprehensive understanding of the molecular composition of these EVs is crucial for unraveling the mechanisms that govern resistance and virulence in C. haemulonii var. vulnera. This study characterizes the proteomic and miRNA-like cargo of EVs from C. haemulonii var. vulnera, revealing components that contribute to its adaptation and survival mechanisms. Proteomic analysis identified 124 EV-specific proteins, including BMH1, TEF1, CDC19, and PDC11, which are linked to processes such as metabolic adaptation, cell wall remodeling, and biofilm formation. miRNA-like molecules associated with mitochondrial function, such as the electron transport chain and regulation of the citric acid cycle, were also detected. These findings provide insights into EV-mediated molecular mechanisms driving fungal pathogenesis and resistance. By characterizing the EV cargo, this study highlights potential targets for antifungal therapies and offers a framework for understanding EV roles in fungal adaptation and pathogenicity.
Importance: The Candida haemulonii complex poses significant clinical challenges due to its intrinsic resistance to conventional antifungal therapies and diagnostic complexities. This research explores the cargo of EVs released by C. haemulonii var. vulnera, revealing a selective mechanism for exporting proteins and RNA molecules critical to the fungus' adaptation and survival in diverse environments. Proteomic and RNA analyses identified molecules involved in metabolic reprogramming, immune modulation, and stress response. These findings highlight the role of EVs in mediating host-pathogen interactions, facilitating immune evasion, and contributing to fungal virulence. Understanding the EV cargo expands our knowledge of fungal biology and underscores the therapeutic potential of targeting EV-associated molecules in antifungal strategies.
Keywords: Candida haemulonii species complex; Candidiasis; Emergent Yeast; Extracellular vesicles.
© 2025 The Authors.
Conflict of interest statement
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
LinkOut - more resources
Full Text Sources
