Removal of dsRNA byproducts using affinity chromatography
- PMID: 40487356
- PMCID: PMC12141893
- DOI: 10.1016/j.omtn.2025.102549
Removal of dsRNA byproducts using affinity chromatography
Abstract
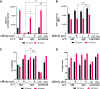
Double-stranded RNA (dsRNA) molecules are immunogenic byproducts of in vitro transcription of single-stranded RNA (ssRNA). Removal of dsRNA from ssRNA is difficult because the byproducts have similar sizes, sequences, and charges to the desired ssRNA. Here, we describe a dsRNA-specific affinity resin that selectively removes dsRNA from ssRNA. Affinity purification reduced dsRNA levels by >100-fold, to as low as ∼0.00007% w/w of total mRNA, with no negative impact on RNA integrity. The purified RNA, synthesized with standard nucleotides, induced no inflammatory response in a reporter cell line assay designed to measure innate immune responses. Purified RNA induced greater protein expression and healthier cells. The immunogenicity of the affinity-purified RNA with standard nucleotides compares favorably to RNA synthesized with modified nucleotides and purified with cellulose or reverse-phase high-performance liquid chromatography (HPLC). dsRNA affinity purification provides a facile and scalable solution to the problem of immunogenic dsRNA byproducts in transcribed RNA. This approach will improve quality and safety of RNA vaccines and therapeutics.
Keywords: MT: Oligonucleotides: Therapies and Applications; RNA vaccines; circular RNA; double-stranded RNA; in vitro transcription; innate immunity; interferon; messenger RNA; self-amplifying RNA; therapeutic RNA.
© 2025 The Author(s).
Conflict of interest statement
All authors are employees of Repligen (N.E.C., M.R.S., R.A.W., K.K., and T.C.S.) or etherna (M.K., S.D.A., J.V.d.H., and S.D.). Repligen manufactures and sells a variety of bioprocessing products, and etherna is an industrial RNA producer.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources

