Efficacy of functional connectome fingerprinting using tangent-space brain networks
- PMID: 40487359
- PMCID: PMC12140576
- DOI: 10.1162/netn_a_00445
Efficacy of functional connectome fingerprinting using tangent-space brain networks
Abstract
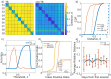
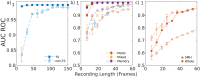
Functional connectomes (FCs) are estimations of brain region interaction derived from brain activity, often obtained from functional magnetic resonance imaging recordings. Quantifying the distance between FCs is important for understanding the relation between behavior, disorders, disease, and changes in connectivity. Recently, tangent space projections, which account for the curvature of the mathematical space of FCs, have been proposed for calculating FC distances. We compare the efficacy of this approach relative to the traditional method in the context of subject identification using the Midnight Scan Club dataset in order to study resting-state and task-based subject discriminability. The tangent space method is found to universally outperform the traditional method. We also focus on the subject identification efficacy of subnetworks. Certain subnetworks are found to outperform others, a dichotomy that largely follows the "control" and "processing" categorization of resting-state networks, and relates subnetwork flexibility with subject discriminability. Identification efficacy is also modulated by tasks, though certain subnetworks appear task independent. The uniquely long recordings of the dataset also allow for explorations of resource requirements for effective subject identification. The tangent space method is found to universally require less data, making it well suited when only short recordings are available.
Keywords: Fingerprinting; Functional connectomes; Resting-state networks.
Plain language summary
Functional connectomes, which describe the similarity between the recorded activity of different brain regions, are ubiquitous for researchers trying to understand brain dynamics on short and long time scales in the presence and absence of neurophysiological diseases. This work applies a tangent space approach, a novel method to calculate the distance between functional connectomes, to a unique high-quality dataset (the Midnight Scan Club) in order to better understand the variability and uniqueness of connectomes across subjects, and how subject identification (also called “fingerprinting”) compares across tasks. We also show that not only does the tangent space method offer greater sensitivity to changes, but it does so with significantly fewer resources, and in some scenarios, reveals qualitatively different interpretations than the traditional method.
© 2025 Massachusetts Institute of Technology.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures



References
-
- Adhikari, M. H., Griffis, J., Siegel, J. S., Thiebaut de Schotten, M., Deco, G., Instabato, A., … Corbetta, M. (2021). Effective connectivity extracts clinically relevant prognostic information from resting state activity in stroke. Brain Communications, 3(4), fcab233. 10.1093/braincomms/fcab233, - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
