Dynamic star allele definitions in Pharmacogenomics: impact on diplotype calls, Phenotype predictions and statin therapy recommendations
- PMID: 40487404
- PMCID: PMC12141247
- DOI: 10.3389/fphar.2025.1584658
Dynamic star allele definitions in Pharmacogenomics: impact on diplotype calls, Phenotype predictions and statin therapy recommendations
Abstract
Introduction: Pharmacogenomics investigates the impact of genetic variation on drug metabolism, enabling personalized medicine through optimized drug selection and dosing. This study examines the effect of the dynamic star allele nomenclature system on diplotypes and therapeutic recommendations using the GeT-RM dataset while also presenting a revised version to address outdated diplotypes.
Materials and methods: PharmVar data up to version 6.2 were downloaded to analyze the evolution of the star allele nomenclature system. FASTQ files from 70 samples of the GeT-RM project were downloaded and aligned to GRCh38, followed by star allele calling using Aldy, PyPGx, and StellarPGx. Diplotypes of the samples were updated based on predefined criteria. Phenotype predictions and therapeutic recommendations were inferred using the PyPGx core API, with CPIC guidelines applied for statin-phenotype combinations.
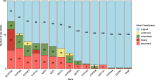
Results: We reevaluated 1400 diplotypes across 20 pharmacogenes in 70 samples from the GeT-RM dataset using three star allele callers: Aldy, PyPGx, and StellarPGx. Our analysis revealed inconsistencies in 15 of 20 pharmacogenes, with 272 (19.4%) diplotypes being outdated. SLCO1B1 showed the highest number of discrepant calls, impacting statin dosing recommendations for NA19226.
Discussion: Our findings demonstrate that outdated allele definitions can alter therapeutic recommendations, emphasizing the need for standardized approaches including mandatory PharmVar version disclosure, implementation of cross-tool validations, and incorporation of confidence metrics for star allele calling tools to ensure reliable pharmacogenomic testing.
Keywords: GeT-RM; diplotype; pharmacogenomics; pharmvar; star allele; statin recommendation.
Copyright © 2025 van der Maas, Denil, Maes, Ertaylan and Volders.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Cooper-DeHoff R. M., Niemi M., Ramsey L. B., Luzum J. A., Tarkiainen E. K., Straka R. J., et al. (2022). enThe clinical pharmacogenetics implementation Consortium guideline for SLCO1B1, ABCG2, and CYP2C9 genotypes and statin-associated musculoskeletal symptoms. Clin. Pharmacol. and Ther. 111, 1007–1021. 10.1002/cpt.2557 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

