Switching from messenger RNAs to noncoding RNAs, METTL3 is a novel colorectal cancer diagnosis and treatment target
- PMID: 40487941
- PMCID: PMC12142237
- DOI: 10.4251/wjgo.v17.i5.104076
Switching from messenger RNAs to noncoding RNAs, METTL3 is a novel colorectal cancer diagnosis and treatment target
Abstract
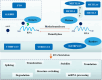
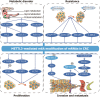
N6-methyladenosine (m6A) modification, one of the most prevalent RNA epigenetic modifications in eukaryotes, constitutes over 60% of all RNA methylation modifications. This dynamic modification regulates RNA processing, maturation, nucleocytoplasmic transport, translation efficiency, phase separation, and stability, thereby linking its dysregulation to diverse physiological and pathological processes. METTL3, a core catalytic component of the methyltransferase complex responsible for m6A deposition, is frequently dysregulated in diseases, including colorectal cancer (CRC). Although METTL3's involvement in CRC pathogenesis has been documented, its precise molecular mechanisms and functional roles remain incompletely understood. METTL3 mediates CRC progression-encompassing proliferation, invasion, drug resistance, and metabolic reprogramming-through m6A-dependent modulation of both coding RNAs and noncoding RNAs. Its regulatory effects are primarily attributed to interactions with key signaling pathways at multiple stages of CRC development. Emerging evidence highlights METTL3 as a promising biomarker for CRC diagnosis and prognosis, as well as a potential therapeutic target. By synthesizing recent advances in METTL3 research within CRC, this review provides critical insights into novel strategies for clinical diagnosis and targeted therapy.
Keywords: Biomarker; Colorectal cancer; Epigenetics; METTL3; N6-methyladenosine; Targeted treatment.
©The Author(s) 2025. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: The authors declare that they have no conflict of interest.
Figures


References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–249. - PubMed
-
- Imamura F, Micha R, Khatibzadeh S, Fahimi S, Shi P, Powles J, Mozaffarian D Global Burden of Diseases Nutrition and Chronic Diseases Expert Group (NutriCoDE) Dietary quality among men and women in 187 countries in 1990 and 2010: a systematic assessment. Lancet Glob Health. 2015;3:e132–e142. - PMC - PubMed
-
- Takayama T, Katsuki S, Takahashi Y, Ohi M, Nojiri S, Sakamaki S, Kato J, Kogawa K, Miyake H, Niitsu Y. Aberrant crypt foci of the colon as precursors of adenoma and cancer. N Engl J Med. 1998;339:1277–1284. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

