Visualizing the transcription and replication of influenza A viral RNAs in cells by multiple direct RNA padlock probing and in situ sequencing (mudRapp-seq)
- PMID: 40488282
- PMCID: PMC12146845
- DOI: 10.1093/nar/gkaf461
Visualizing the transcription and replication of influenza A viral RNAs in cells by multiple direct RNA padlock probing and in situ sequencing (mudRapp-seq)
Abstract
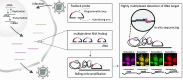
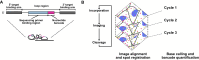
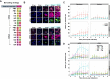
Influenza A viruses (IAVs) contain eight negative-sense single-stranded viral RNA (vRNA) molecules, which are transcribed into messenger RNA (mRNA) and replicated via complementary RNA (cRNA). These processes are tightly regulated, but the precise molecular mechanisms governing the switch from transcription to replication remain elusive. Here, we introduce multiple direct RNA-assisted padlock probing in combination with in situ sequencing (mudRapp-seq) to visualize the transcription and replication of all eight IAV vRNA and mRNA molecules at the single-cell level. We demonstrate that direct RNA padlock probing is three times more efficient than conventional probes that target cDNA. Individual probes showed variations in efficiency, partly due to the RNA structure of the target, which was mitigated by employing multiple padlock probes per target. Applying mudRapp-seq to an infection time course, we observed early mRNA expression, followed by vRNA accumulation ∼3 h later. Individual viral segments exhibited differential expression, particularly in the mRNA population. Both bulk and single-cell analyses revealed a correlation between the expression of "M" mRNA and the onset of the transcription-to-replication switch. Our findings demonstrate that mudRapp-seq offers significant potential for elucidating viral replication mechanisms and may be applicable to studying other RNA viruses and cellular RNA processes.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

