Deciphering meiotic chromatin organization by SYCP3
- PMID: 40488283
- PMCID: PMC12146848
- DOI: 10.1093/nar/gkaf460
Deciphering meiotic chromatin organization by SYCP3
Abstract
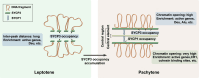
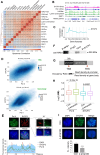
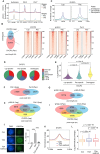
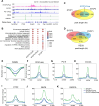
Chromatin structure during meiosis is different from somatic cells due to the assembly of the synaptonemal complex between homologous chromosome axes. However, genome-wide organizing principles of this meiosis-specific multiprotein complex remain mysterious despite intensive super-resolution imaging analysis. Here, we profiled chromatin occupancy of SYCP3, the key chromatin organizer of synaptonemal complex, in mouse spermatocytes, and showed its enrichment at open chromatin regions. Moreover, SYCP3 occupancy was largely inherited from the leptotene to pachytene stage, facilitated by transcription and fibrous assembly, and was enriched at specific SINE repeats. We also identified SYCP1-occupied regions mainly as a subpopulation of SYCP3-occupied regions with high cohesin enrichment. Collectively, our results demonstrate genome-wide profiling of SYCP3 in mouse meiosis and reveal that its occupancy is a dynamic process modulated by chromatin-related events.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

