Metabolism and response to stress gene signatures reveal ulcerative colitis heterogeneity and identify patients with increased response to therapy
- PMID: 40488582
- PMCID: PMC12203003
- DOI: 10.1093/ecco-jcc/jjaf092
Metabolism and response to stress gene signatures reveal ulcerative colitis heterogeneity and identify patients with increased response to therapy
Abstract
Background and aims: Ulcerative colitis (UC) therapies lead to variable remission and response rates in patients participating in clinical trials, likely due to interindividual target variability, differences in active biological pathways, feedback, and/or resistance mechanisms. Here, we stratified patients into subtypes by characterizing heterogeneity using mucosal biopsy transcriptomics data.
Methods: Transcriptomics data from an andecaliximab phase 2/3 study in patients with UC were scored for gene signature enrichment. Eleven Reactome gene sets, moderately correlated with histological disease activity using Robarts Histopathology Index with low correlation to each other, were selected and evaluated in baseline gene expression data of ustekinumab, infliximab, and adalimumab clinical trials in patients with UC.
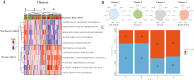
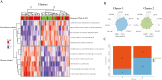
Results: Of 11 gene sets, referred to as "Metabolism and Response to Stress" (MARS) signatures, 5 correlated with "non-disease" mucosa and 6 with "disease-related" mucosa. Clustering baseline andecaliximab samples scored with MARS revealed 3 clusters with low non-disease/high disease-related, high non-disease/low disease-related, or a mixture. Importantly, these clusters did not correlate with patient demographics, clinical characteristics, or disease activity metrics. Clustering baseline data from other clinical trials (anti-interleukin-12/23 and anti-tumor necrosis factor) in patients with UC scored with MARS showed that patients in low non-disease/high disease-related baseline score clusters less likely to achieve treatment response.
Conclusions: We identified and evaluated a novel, multi-dimensional signature gene set to characterize previously undefined heterogeneity in patients with UC and identify patients less likely to respond to therapy. This approach offers potential utility to define clinical trial populations, enrich for clinical responders, and identify difficult-to-treat populations for therapeutic development.
Keywords: biomarkers; clinical trials; heterogeneity.
© The Author(s) 2025. Published by Oxford University Press on behalf of European Crohn’s and Colitis Organisation.
Conflict of interest statement
B.L. is a former employee of Alimentiv Inc. and is currently an employee of AnaptysBio Inc.
M.F., B.S., M.I.S., and W.T. are employees of Alimentiv Inc.
V.J. has received consulting/advisory board fees from AbbVie, Alimentiv Inc., Arena Pharmaceuticals, Asahi Kasei Pharma, Asieris, Astra Zeneca, Bristol Myers Squibb, Celltrion, Eli Lilly, Ferring, Flagship Pioneering, Fresenius Kabi, Galapagos, GlaxoSmithKline, Genentech, Gilead, Janssen, Merck, Mylan, Pandion, Pendopharm, Pfizer, Protagonist, Prometheus, Reistone Biopharma, Roche, Sandoz, Second Genome, Takeda, Teva, TopiVert, Ventyx, and Vividion; and has received speaker’s fees from AbbVie, Ferring, Galapagos, Janssen Pfizer Shire, Takeda, and Fresenius Kabi.
C.M. has received consulting fees from AbbVie, Alimentiv Inc., Amgen, AVIR Pharma, Bristol Myers Squibb, Ferring, Fresenius Kabi, Janssen, McKesson, Mylan, Takeda, Pfizer, and Roche; has received speaker’s fees from AbbVie, Amgen, AVIR Pharma Inc., Alimentiv Inc., Ferring, Janssen, Takeda, and Pfizer; and has received research support from Pfizer.
N.V.C. is a former employee of AcelaBio, Inc.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

