Shared hub genes in membranous nephropathy and kidney renal clear cell carcinoma: investigating molecular overlap and tumor progression
- PMID: 40489038
- PMCID: PMC12149382
- DOI: 10.1007/s12672-025-02701-1
Shared hub genes in membranous nephropathy and kidney renal clear cell carcinoma: investigating molecular overlap and tumor progression
Abstract
Background: Membranous nephropathy (MN) and kidney renal clear cell carcinoma (KIRC) are distinct kidney diseases with potential shared molecular mechanisms. Identifying common biomarkers may improve our understanding of disease pathogenesis and provide novel diagnostic and therapeutic targets.
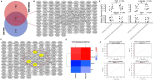
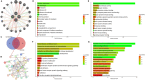
Methods: The study primarily employed bioinformatics tools to analyze publicly available datasets to identify differentially expressed genes (DEGs) and hub genes in KIRC and MN. Functional interactions of the common DEGs were explored using protein-protein interaction (PPI) networks, and hub genes were further investigated through gene expression databases such as GSCA and UALCAN. Gene Set Enrichment Analysis (GSEA) was used to assess functional enrichment and tumor-driving potential. These bioinformatic results were then experimentally validated by knocking down FYN and LGALS8 in 786-O cells using siRNA, followed by RT-qPCR, protein analysis, and functional assays.
Results: The study identified four hub genes (FYN, LGALS8, MAGI2, and WT1) in KIRC and MN, with FYN and LGALS8 upregulated and MAGI2 and WT1 downregulated. Bioinformatics validation showed excellent diagnostic performance and confirmed methylation and mutation patterns. Higher FYN and LGALS8 expression were linked to poorer survival. miRNA downregulation was validated in KIRC cell lines. Functional analysis revealed that FYN and LGALS8 promote KIRC progression through the ErbB signaling pathway, and knockdown experiments reduced cell proliferation, migration, and colony formation.
Conclusion: Our findings identify FYN, LGALS8, MAGI2, and WT1 as hub genes in KIRC, with potential diagnostic and prognostic value. These genes play significant roles in methylation, mutation, and immune regulation in KIRC. However, the results from the limited MN samples suggest possible roles of these genes in MN pathology, but further studies are required to fully assess the relevance of these findings to MN.
Keywords: ErbB signaling pathway; FYN; KIRC; LGALS8; MN.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests. Clinical trial number: Not applicable.
Figures







Similar articles
-
The discovery of promising candidate biomarkers in kidney renal clear cell carcinoma: evidence from the in-depth analysis of high-throughput data.Am J Cancer Res. 2023 Sep 15;13(9):4288-4304. eCollection 2023. Am J Cancer Res. 2023. PMID: 37818073 Free PMC article.
-
The promising novel biomarkers and candidate small molecule drugs in kidney renal clear cell carcinoma: Evidence from bioinformatics analysis of high-throughput data.Mol Genet Genomic Med. 2019 May;7(5):e607. doi: 10.1002/mgg3.607. Epub 2019 Feb 21. Mol Genet Genomic Med. 2019. PMID: 30793530 Free PMC article.
-
Deciphering key genes involved in cisplatin resistance in kidney renal clear cell carcinoma through a combined in silico and in vitro approach.Oncol Res. 2023 Sep 15;31(6):899-916. doi: 10.32604/or.2023.030760. eCollection 2023. Oncol Res. 2023. PMID: 37744271 Free PMC article.
-
In vitro analysis of PI3K pathway activation genes for exploring novel biomarkers and therapeutic targets in clear cell renal carcinoma.Am J Transl Res. 2023 Jul 15;15(7):4851-4872. eCollection 2023. Am J Transl Res. 2023. PMID: 37560222 Free PMC article.
-
Identifying and validating MMP family members (MMP2, MMP9, MMP12, and MMP16) as therapeutic targets and biomarkers in kidney renal clear cell carcinoma (KIRC).Oncol Res. 2024 Mar 20;32(4):737-752. doi: 10.32604/or.2023.042925. eCollection 2024. Oncol Res. 2024. PMID: 38560573 Free PMC article.
References
-
- Reinhard L, Stahl RA, Hoxha E. Is primary membranous nephropathy a complement mediated disease? Mol Immunol. 2020;128:195–204. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
