Changes of respiratory microbiota associated with prognosis in pulmonary infection patients with invasive mechanical ventilation-supported respiratory failure
- PMID: 40489326
- PMCID: PMC12150602
- DOI: 10.1080/07853890.2025.2514093
Changes of respiratory microbiota associated with prognosis in pulmonary infection patients with invasive mechanical ventilation-supported respiratory failure
Abstract
Background: Respiratory failure (RF) is an important cause of intensive care unit (ICU) admission and mortality due to respiratory diseases. This study aimed to evaluate the clinical performance of metagenomic next-generation sequencing (mNGS) testing in pathogen diagnosis, medication guidance and to explore dynamic changes in the respiratory microbiota associated with prognosis.
Methods: This multicenter retrospective study enrolled ICU patients from five hospitals who underwent invasive mechanical ventilation (IMV) and had pathogenic microorganisms identified by both mNGS and conventional microbiological tests (CMT) from December 2021 to April 2024. Patients were classified into two groups based on discharge outcomes: survivors (n=122) and non-survivors (n=35).
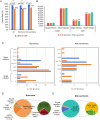
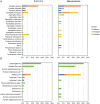
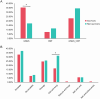
Results: Compared with the survivors, non-survivors had a significantly higher proportion of smokers, dyspnea, type I RF, blood urea nitrogen, and C-reactive protein (p < 0.05). All the above indicators were identified as independent risk factors for mortality, except for type I RF. mNGS showed a better performance for pathogen identification than CMT in both groups, and nearly 60% showed consistent results between the two methods. Among survivors, antibiotic adjustment was mainly based on mNGS results (35.25%), whereas non-survivors more frequently received adjustments based on mNGS and CMT results (34.29%). The richness and abundance of lung microorganisms in the non-survivors were significantly lower than those in the survivors (p < 0.05).
Conclusions: mNGS is a promising method for identifying pathogens in pulmonary infections in IMV-supported RF patients and for exploring changes in lung microbial composition to provide a reference for patient prognosis.
Keywords: Invasive mechanical ventilation; lung microbial; metagenomic next-generation sequencing; pulmonary infection; respiratory failure.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Mechanical ventilation for newborn infants with respiratory failure due to pulmonary disease.Cochrane Database Syst Rev. 2002;2002(4):CD002770. doi: 10.1002/14651858.CD002770. Cochrane Database Syst Rev. 2002. PMID: 12519575 Free PMC article.
-
The clinical utility of bronchoalveolar lavage fluid metagenomic next-generation sequencing in immunocompromised critically ill patients with invasive pulmonary aspergillosis: a multicenter retrospective study.Microbiol Spectr. 2025 Aug 5;13(8):e0058425. doi: 10.1128/spectrum.00584-25. Epub 2025 Jun 12. Microbiol Spectr. 2025. PMID: 40503829 Free PMC article.
-
Non-invasive ventilation for the management of acute hypercapnic respiratory failure due to exacerbation of chronic obstructive pulmonary disease.Cochrane Database Syst Rev. 2017 Jul 13;7(7):CD004104. doi: 10.1002/14651858.CD004104.pub4. Cochrane Database Syst Rev. 2017. PMID: 28702957 Free PMC article.
-
Performance of broad-spectrum targeted next-generation sequencing in lower respiratory tract infections in ICU patients: a prospective observational study.Crit Care. 2025 Jun 4;29(1):226. doi: 10.1186/s13054-025-05470-z. Crit Care. 2025. PMID: 40468429 Free PMC article.
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
