Multiple introductions of equine influenza virus into the United Kingdom resulted in widespread outbreaks and lineage replacement
- PMID: 40489557
- PMCID: PMC12236680
- DOI: 10.1371/journal.ppat.1013227
Multiple introductions of equine influenza virus into the United Kingdom resulted in widespread outbreaks and lineage replacement
Abstract
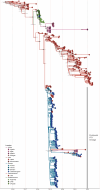
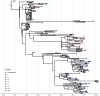
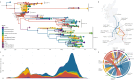
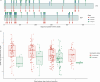
Influenza A viruses (IAVs) are prime examples of emerging viruses in humans and animals. IAV circulation in domestic animals poses a pandemic risk as it provides new opportunities for zoonotic infections. The recent emergence of H5N1 IAV in cows and subsequent spread over multiple states within the USA, together with reports of spillover infections in humans, cats and mice highlight this issue. The horse is a domestic animal in which an avian-origin IAV lineage has been circulating for >60 years. In 2018/19, a Florida Clade 1 (FC1) virus triggered one of the largest epizootics recorded in the UK, which led to the replacement of the Equine Influenza Virus (EIV) Florida Clade 2 (FC2) lineage that had been circulating in the country since 2003. We integrated geographical, epidemiological, and virus genetic data to determine the virological and ecological factors leading to this epizootic. By combining newly-sequenced EIV complete genomes derived from UK outbreaks with existing genomic and epidemiological information, we reconstructed the nationwide viral spread and analysed the global evolution of EIV. We show that there was a single EIV FC1 introduction from the USA into Europe, and multiple independent virus introductions from Europe to the UK. At the UK level, three English regions (East, West Midlands, and North-West) were the main sources of virus during the epizootic, and the number of affected premises together with the number of horses in the local area were found as key predictors of viral spread within the country. At the global level, phylogeographic analysis evidenced a source-sink model for intercontinental EIV migration, with a source population evolving in the USA and directly or indirectly seeding viral lineages into sink populations in other continents. Our results provide insight on the underlying factors that influence IAV spread in domestic animals.
Copyright: © 2025 Mojsiejczuk et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

