Imprints of somatic hypermutation on B-cell receptor immunoglobulins post-infection versus post-vaccination against SARS-CoV-2
- PMID: 40489958
- PMCID: PMC12148301
- DOI: 10.1093/immhor/vlaf021
Imprints of somatic hypermutation on B-cell receptor immunoglobulins post-infection versus post-vaccination against SARS-CoV-2
Abstract
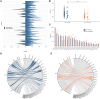
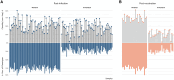
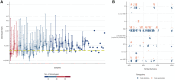
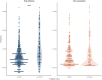
Published evidence supports significant heterogeneity of immune responses among individuals infected with or vaccinated against SARS-CoV-2. This highlights the need for in-depth investigation of the implicated processes toward refined understanding and improved management of COVID-19. The main objective of the present study was to investigate the dynamics of B cell responses to SARS-CoV-2, focusing on how initial infection and subsequent vaccination influence the immunoglobulin gene repertoire, with special emphasis on the impact of somatic hypermutation (SHM) on antibody maturation. Samples were collected from 81 individuals infected by SARS-CoV-2 in the municipality of Vo' during the first pandemic wave in 2020. For 25 of them, sampling was repeated 7 d after completing the primary vaccination series. Deep immunogenetic analysis of the B-cell receptor immunoglobulin (BcR IG) gene repertoire was performed using targeted next-generation sequencing. Bioinformatics analysis focused on repertoire metrics, prediction of IG antigen specificity, and detailed profiling of the SHM patterns. Significant expansions of unmutated sequences early post-infection suggest extrafollicular B cell maturation. In contrast, vaccination promoted SHM acquisition, indicating a germinal center-dependent response, and pronounced repertoire renewal. Restricted SHMs in SARS-homologous clonotypes along with preferential targeting of specific codons within the VH domain post-vaccination support ongoing affinity maturation within germinal centers. Differences in the BcR IG profiles post-infection versus post-vaccination allude to distinct trajectories in B cell maturation. Distinct profiles of SHM targeting reflect ongoing affinity maturation post-vaccination, with implications for optimizing preventive and therapeutic interventions against COVID-19.
Keywords: B-cell receptor gene repertoire; COVID-19; adaptive immunity; somatic hypermutation.
© The Author(s) 2025. Published by Oxford University Press on behalf of The American Association of Immunologists.
Conflict of interest statement
The authors declare no relevant conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

