microRNA-1 regulates metabolic flexibility by programming adult skeletal muscle pyruvate metabolism
- PMID: 40490136
- PMCID: PMC12214129
- DOI: 10.1016/j.molmet.2025.102182
microRNA-1 regulates metabolic flexibility by programming adult skeletal muscle pyruvate metabolism
Erratum in
-
Corrigendum to "microRNA-1 regulates metabolic flexibility by programming skeletal muscle pyruvate metabolism" [Mol Metabol 98 (2025) 1-23/102182].Mol Metab. 2025 Oct;100:102223. doi: 10.1016/j.molmet.2025.102223. Epub 2025 Aug 7. Mol Metab. 2025. PMID: 40774330 Free PMC article. No abstract available.
Abstract
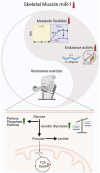
Objective: Metabolic flexibility refers to the ability of tissues to adjust cellular fuel choice in response to conditional changes in metabolic demand and activity. A loss of metabolic flexibility is a defining feature of various diseases and cellular dysfunction. This study investigated the role of microRNA-1 (miR-1), the most abundant microRNA in skeletal muscle, in maintaining whole-body metabolic flexibility.
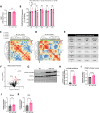
Methods: We used an inducible, skeletal muscle-specific knockout (KO) mouse model to examine miR-1 function. Argonaute 2 enhanced crosslinking and immunoprecipitation sequencing (AGO2 eCLIP-seq) and RNA-seq analyses identified miR-1 target genes. Metabolism was investigated using metabolomics, proteomics, and comprehensive bioenergetic and activity phenotyping. Corroborating information was provided from cell culture, C. elegans, and exercised human muscle tissue.
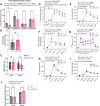
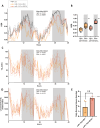
Results: miR-1 KO mice demonstrated loss of diurnal oscillations in whole-body respiratory exchange ratio and higher fasting blood glucose. For the first time, we identified bona fide miR-1 target genes in adult skeletal muscle that regulated pyruvate metabolism through mechanisms including the alternative splicing of pyruvate kinase (Pkm). The maintenance of metabolic flexibility by miR-1 was necessary for sustained endurance activity in mice and in C. elegans. Loss of metabolic flexibility in the miR-1 KO mouse was rescued by pharmacological inhibition of the miR-1 target, monocarboxylate transporter 4 (MCT4), which redirects glycolytic carbon flux toward oxidation. The physiological down-regulation of miR-1 in response to hypertrophic stimuli caused a similar metabolic reprogramming necessary for muscle cell growth.
Conclusions: These data identify a novel post-transcriptional mechanism of whole-body metabolism regulation mediated by a tissue-specific miRNA.
Keywords: Aerobic glycolysis; MCT4; PKM; Resistance training; VB124; eCLIP-seq.
Copyright © 2025 The Author(s). Published by Elsevier GmbH.. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





Update of
-
microRNA-1 Regulates Metabolic Flexibility in Skeletal Muscle via Pyruvate Metabolism.bioRxiv [Preprint]. 2024 Aug 10:2024.08.09.607377. doi: 10.1101/2024.08.09.607377. bioRxiv. 2024. Update in: Mol Metab. 2025 Aug;98:102182. doi: 10.1016/j.molmet.2025.102182. PMID: 39149347 Free PMC article. Updated. Preprint.
References
-
- Nielsen M., Hansen J.H., Hedegaard J., Nielsen R.O., Panitz F., Bendixen C., et al. MicroRNA identity and abundance in porcine skeletal muscles determined by deep sequencing. Anim Genet. 2010;41(2):159–168. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

