Regulated microexon alternative splicing in single neurons tunes synaptic function
- PMID: 40490601
- PMCID: PMC12287369
- DOI: 10.1038/s44319-025-00493-7
Regulated microexon alternative splicing in single neurons tunes synaptic function
Abstract
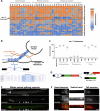
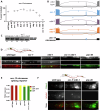
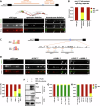
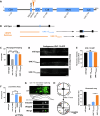
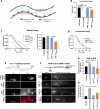
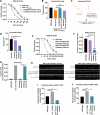
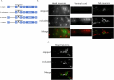
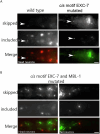
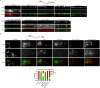
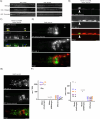
Microexons are important components of the neuronal transcriptome. Though tiny, their splicing is essential for neuronal development and function. Microexons are typically included in the nervous system and skipped in other tissues, but less is known about whether they are alternatively spliced across neuron types, and if so what the regulatory mechanisms and functional consequences might be. We set out to globally address this question in C. elegans using deep single-cell transcriptomes and in vivo splicing reporters. We find widespread alternative microexon splicing across neuron types. Focusing on a broadly-conserved 9-nucleotide exon in the synaptic vesicle gene unc-13, we find that it is completely skipped in olfactory neurons, but completely included in motor neurons. This splicing pattern is established by two neuronal RNA binding proteins which recruit spliceosomal component PRP-40 to mediate microexon inclusion. Cell-specific microexon alternative splicing is functionally important, as forcing microexon inclusion causes olfactory defects, while forcing microexon skipping causes locomotory defects. These locomotory defects are caused by decreased inhibitory motor neuron synaptic transmission and altered synaptic vesicle distribution. Regulatory features of unc-13 microexon splicing are broadly conserved: related MUN-domain genes in worms, flies, and mice invariably encode microexons, and those we tested are subject to similar regulatory principles (e.g. included in motor neurons, skipped in olfactory neurons, and regulated by the same two RNA binding proteins). Thus, not only is microexon inclusion important for nervous system function, but microexon alternative splicing across neurons is important for tuning neuronal function in individual cell types.
Keywords: unc-13; Alternative Splicing; Microexon; Neuron; Splicing.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures


Similar articles
-
Conserved role for spliceosomal component PRPF40A in microexon splicing.RNA. 2024 Dec 16;31(1):43-50. doi: 10.1261/rna.080142.124. RNA. 2024. PMID: 39389624
-
Spliceosomal component PRP-40 is a central regulator of microexon splicing.Cell Rep. 2021 Aug 3;36(5):109464. doi: 10.1016/j.celrep.2021.109464. Cell Rep. 2021. PMID: 34348142 Free PMC article.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Fabricating mice and dementia: opening up relations in multi-species research.In: Jenkins N, Jack-Waugh A, Ritchie L, editors. Multi-Species Dementia Studies. Bristol (UK): Bristol University Press; 2025 Feb 25. Chapter 2. In: Jenkins N, Jack-Waugh A, Ritchie L, editors. Multi-Species Dementia Studies. Bristol (UK): Bristol University Press; 2025 Feb 25. Chapter 2. PMID: 40690569 Free Books & Documents. Review.
References
-
- Bargmann CI, Hartwieg E, Horvitz HR (1993) Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74:515–527 - PubMed
-
- Barrett A, Varol E, Weinreb A, Taylor SR, McWhirter RM, Cros C, Vidal B, Basavaraju M, Poff A, Tipps JA et al (2025) Integrating bulk and single cell RNA-seq refines transcriptomic profiles of individual C. elegans neurons. eLife 14:RP106183
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

