Alternative Splicing and CaV-Associated Channelopathies
- PMID: 40490926
- PMCID: PMC12149502
- DOI: 10.1002/wrna.70016
Alternative Splicing and CaV-Associated Channelopathies
Abstract
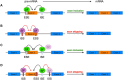
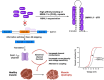
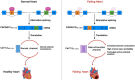
Voltage-gated calcium channels (VGCCs) are multi-subunit ion channel proteins that control and regulate a wide array of physiological processes. Their dysfunction has been implicated in several neurological, cardiac, psychiatric, endocrine, oncogenic, and muscular disorders. The diverse and specialized cellular functions involving VGCC-mediated calcium signaling stem from two primary mechanisms: differential and cell-specific expression of pore-forming (α1) and auxiliary subunit genes, and extensive alternative splicing of their pre-mRNA. All the 10 α1-encoding genes undergo alternative splicing to generate a wide array of cell-specific CaV variants with distinct biophysical, pharmacological, and protein-protein interaction properties. This proteomic diversity and the associated cell-specific expression signature of CaV splice variants are tightly regulated by trans-acting splicing factors-RNA-binding proteins that control the inclusion or skipping of alternatively spliced exons during post-transcriptional pre-mRNA processing. The discovery that several channelopathies are caused by aberrant splicing due to genetic mutations in either cis-acting binding elements on the pre-mRNA or in core splicing machinery components highlights the crucial role of alternative splicing in VGCC-related pathologies. These insights have opened new therapeutic avenues, as targeting the alternative splicing of disease-associated specific exons has recently emerged as a novel, promising treatment for neurodevelopmental disorders and channelopathies associated with splicing dysfunction.
Keywords: alternative splicing; calcium channel; disease; ion channel; therapy.
© 2025 The Author(s). WIREs RNA published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

