Improvement of D-lactic acid production from methanol by metabolically engineered Komagataella phaffii via ultra-violet mutagenesis
- PMID: 40491503
- PMCID: PMC12148619
- DOI: 10.1016/j.mec.2025.e00262
Improvement of D-lactic acid production from methanol by metabolically engineered Komagataella phaffii via ultra-violet mutagenesis
Abstract
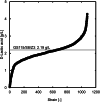
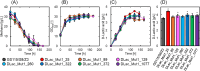
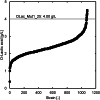
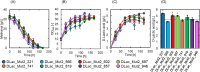
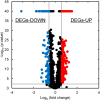
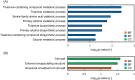
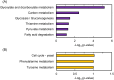
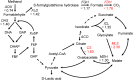
Methanol has attracted attention as an alternative carbon source to petroleum. Komagataella phaffii, a methanol-assimilating yeast, is a useful host for the chemical production from methanol. A previous study successfully constructed a metabolically engineered K. phaffii GS115/S8/Z3 strain capable of producing D-lactic acid from methanol. In this study, we aimed to develop a strain with improved D-lactic acid production by applying ultra-violet mutagenesis to the D-lactic acid-producing strain, GS115/S8/Z3. The resulting mutant strain DLac_Mut2_221 produced 5.38 g/L of D-lactic acid from methanol, a 1.52-fold increase compared to the parent strain GS115/S8/Z3. The transcriptome analysis of the mutant DLac_Mut2_221 identified 158 differentially expressed genes, providing insights into key mechanisms contributing to enhanced D-lactic acid production. Metabolic engineering strategies for K. phaffii based on the knowledge gained from this study will contribute to improving the productivity of various useful chemicals from methanol.
Keywords: D-lactic acid; Komagataella phaffii; Methanol; Transcriptome analysis; Ultra-violet mutagenesis; Yeast.
© 2025 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
Enhancing D-lactic acid production by optimizing the expression of D-LDH gene in methylotrophic yeast Komagataella phaffii.Biotechnol Biofuels Bioprod. 2024 Dec 22;17(1):149. doi: 10.1186/s13068-024-02596-0. Biotechnol Biofuels Bioprod. 2024. PMID: 39710696 Free PMC article.
-
Toward the construction of a technology platform for chemicals production from methanol: D-lactic acid production from methanol by an engineered yeast Pichia pastoris.World J Microbiol Biotechnol. 2019 Feb 4;35(2):37. doi: 10.1007/s11274-019-2610-4. World J Microbiol Biotechnol. 2019. PMID: 30715602
-
Overproduction of Patchoulol in Metabolically Engineered Komagataella phaffii.J Agric Food Chem. 2023 Feb 1;71(4):2049-2058. doi: 10.1021/acs.jafc.2c08228. Epub 2023 Jan 22. J Agric Food Chem. 2023. PMID: 36681940
-
What makes Komagataella phaffii non-conventional?FEMS Yeast Res. 2021 Dec 24;21(8):foab059. doi: 10.1093/femsyr/foab059. FEMS Yeast Res. 2021. PMID: 34849756 Free PMC article. Review.
-
Komagataella phaffii as Emerging Model Organism in Fundamental Research.Front Microbiol. 2021 Jan 11;11:607028. doi: 10.3389/fmicb.2020.607028. eCollection 2020. Front Microbiol. 2021. PMID: 33505376 Free PMC article. Review.
References
-
- Burg J.M., Cooper C.B., Ye Z., Reed B.R., Moreb E.A., Lynch M.D. Large-scale bioprocess competitiveness: the potential of dynamic metabolic control in two-stage fermentations. Curr. Opin. Chem. Eng. 2016;14:121–136. doi: 10.1016/j.coche.2016.09.008. - DOI
LinkOut - more resources
Full Text Sources

