Uncovering codon usage patterns during murine embryogenesis and tissue-specific developmental diseases
- PMID: 40491567
- PMCID: PMC12146342
- DOI: 10.3389/fgene.2025.1554773
Uncovering codon usage patterns during murine embryogenesis and tissue-specific developmental diseases
Abstract
Introduction: Mouse models share significant genetic similarities with humans and have expanded our understanding of how embryonic tissue-specific genes influence disease states. By improved analyses of temporal, transcriptional data from these models, we can capture unique tissue codon usage patterns and determine how deviations from these patterns can influence developmental disorders.
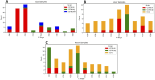
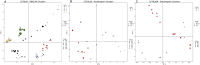
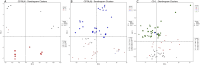
Methods: We analyzed transcriptomic-weighted data from four mouse strains across three different germ layer tissues (liver, heart, and eye) and through embryonic stages. Applying a multifaceted approach, we calculated relative synonymous codon usage, reduced the dimensionality, and employed machine learning clustering techniques.
Results and discussion: These techniques identified relative synonymous codon usage differences/similarities among strains and deviations in codon usage patterns between healthy and disease-linked genes. Original transcriptomic mouse data and RefSeq gene sequences can be found at the associated Mouse Embryo CoCoPUTs (codon and codon pair usage tables) website. Future studies can leverage this resource to uncover further insights into the dynamics of embryonic development and the corresponding codon usage biases that are paramount to understanding disease processes of embryologic origin.
Keywords: clustering methods; disease-associated comparison; machine learning; mouse embryology; relative synonymous codon usage; tissue-specific; transcriptomic-weighted.
Copyright © 2025 Fumagalli, Smith, Lin, Paul, Campbell, Santana-Quintero, Golikov, Ibla, Bar, Komar, Hunt, DiCuccio and Kimchi-Sarfaty.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Mouse embryo CoCoPUTs: novel murine transcriptomic-weighted usage website featuring multiple strains, tissues, and stages.BMC Bioinformatics. 2024 Sep 6;25(1):294. doi: 10.1186/s12859-024-05906-3. BMC Bioinformatics. 2024. PMID: 39242990 Free PMC article.
-
Distinct signatures of codon and codon pair usage in 32 primary tumor types in the novel database CancerCoCoPUTs for cancer-specific codon usage.Genome Med. 2021 Jul 28;13(1):122. doi: 10.1186/s13073-021-00935-6. Genome Med. 2021. PMID: 34321100 Free PMC article.
-
Developmental stage related patterns of codon usage and genomic GC content: searching for evolutionary fingerprints with models of stem cell differentiation.Genome Biol. 2007;8(3):R35. doi: 10.1186/gb-2007-8-3-r35. Genome Biol. 2007. PMID: 17349061 Free PMC article.
-
Codon usage bias.Mol Biol Rep. 2022 Jan;49(1):539-565. doi: 10.1007/s11033-021-06749-4. Epub 2021 Nov 25. Mol Biol Rep. 2022. PMID: 34822069 Free PMC article. Review.
-
Synonymous but Not Silent: The Codon Usage Code for Gene Expression and Protein Folding.Annu Rev Biochem. 2021 Jun 20;90:375-401. doi: 10.1146/annurev-biochem-071320-112701. Epub 2021 Jan 13. Annu Rev Biochem. 2021. PMID: 33441035 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

