This is a preprint.
Understanding Plasmodium vivax recurrent infections using an amplicon deep sequencing assay, PvAmpSeq, identity-by-descent and model-based classification
- PMID: 40492072
- PMCID: PMC12148277
- DOI: 10.1101/2025.05.26.25327775
Understanding Plasmodium vivax recurrent infections using an amplicon deep sequencing assay, PvAmpSeq, identity-by-descent and model-based classification
Abstract
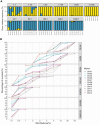
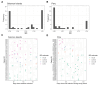
Plasmodium vivax infections are characterised by recurrent bouts of blood-stage parasitaemia. Understanding the genetic relatedness of recurrences can distinguish whether these are caused by relapse, reinfection, or recrudescence, which is critical to understand treatment efficacy and transmission dynamics. We developed PvAmpseq, an amplicon sequencing assay targeting 11 SNP-rich regions of the P. vivax genome. PvAmpSeq was validated on field isolates from a clinical trial in the Solomon Islands and a longitudinal observational cohort in Peru, and statistical models were applied for genetic classification of infection pairs. In the Solomon Islands trial, where participants received antimalarials at baseline, half of the recurrent infections were caused by parasites with >50% relatedness to the baseline infection, with statistical models classifying 25% and 25% as probable relapses and recrudescences, respectively. In the Peruvian cohort, 26% of recurrences were likely relapses. PvAmpSeq provides high-resolution genotyping to characterise P. vivax recurrences, offering insights into transmission and treatment outcomes.
Keywords: Plasmodium vivax; genetic diversity; microhaplotypes; recurrences; relapses.
Conflict of interest statement
Declaration of interest The authors declare no competing interests.
Figures



Similar articles
-
Artemisinin-based combination therapy for treating uncomplicated Plasmodium vivax malaria.Cochrane Database Syst Rev. 2013 Oct 25;2013(10):CD008492. doi: 10.1002/14651858.CD008492.pub3. Cochrane Database Syst Rev. 2013. PMID: 24163021 Free PMC article.
-
Microhaplotype deep sequencing assays to capture Plasmodium vivax infection lineages.Nat Commun. 2025 Aug 5;16(1):7192. doi: 10.1038/s41467-025-62357-x. Nat Commun. 2025. PMID: 40764298 Free PMC article. Clinical Trial.
-
Mass drug administration for malaria.Cochrane Database Syst Rev. 2021 Sep 29;9(9):CD008846. doi: 10.1002/14651858.CD008846.pub3. Cochrane Database Syst Rev. 2021. PMID: 34585740 Free PMC article.
-
Primaquine or other 8-aminoquinoline for reducing P. falciparum transmission.Cochrane Database Syst Rev. 2014 Jun 30;(6):CD008152. doi: 10.1002/14651858.CD008152.pub3. Cochrane Database Syst Rev. 2014. Update in: Cochrane Database Syst Rev. 2015 Feb 19;(2):CD008152. doi: 10.1002/14651858.CD008152.pub4. PMID: 24979199 Free PMC article. Updated.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
References
-
- World Health Organisation (2024). World malaria report 2024.
-
- Robinson L.J., Wampfler R., Betuela I., Karl S., White M.T., Li Wai Suen C.S., Hofmann N.E., Kinboro B., Waltmann A., Brewster J., et al. (2015). Strategies for understanding and reducing the Plasmodium vivax and Plasmodium ovale hypnozoite reservoir in Papua New Guinean children: a randomised placebo-controlled trial and mathematical model. PLoS Med. 12, e1001891. - PMC - PubMed
-
- World Health Organisation (2021). Informal consultation on methodology to distinguish reinfection from recrudescence in high malaria transmission areas.
-
- World Health Organisation (2008). Methods and techniques for clinical trials on antimalarial drug efficacy: genotyping to identify parasite populations.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
