Ccdc13 is essential for the assembly of ciliary central microtubules
- PMID: 40492112
- PMCID: PMC12147715
- DOI: 10.1093/nsr/nwaf095
Ccdc13 is essential for the assembly of ciliary central microtubules
Abstract
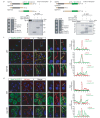
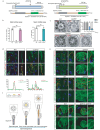
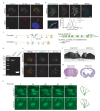
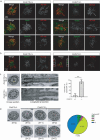
Motile cilia are critical for diverse cellular activities, affecting the survival and development of most eukaryotic organisms. Central microtubules (MTs), which are located in the lumen of ciliary axonemes, are non-centrosomal MTs that are crucial for motile cilia beating. However, the formation mechanism of central MTs remains elusive. Here, by using a Drosophila model, we identify Ccdc13 as a novel regulator for the assembly of central MTs. We show that Ccdc13 localizes along the central MTs and is essential for its formation in sperm flagella, with its deletion consequently affecting the sperm motility and the fertility of male flies. Mechanistically, we demonstrated that Ccdc13 directly interacts with Spef1, acting upstream of Spef1 to regulate central MT elongation. Remarkably, we demonstrated that the role of Ccdc13 in ciliary central MT formation is conserved in mammals. Ccdc13 deficiency in mice leads to the loss of central MTs in motile ependymal cilia, resulting in abnormal cilia motility and hydrocephalus. Our results mark the discovery of Ccdc13 as a novel regulator for ciliary central MT assembly and reveal that the Ccdc13-Spef1 complex is an evolutionarily conserved module that is critical for central MT formation in motile cilia of both flies and mammals.
Keywords: Ccdc13; Drosophila; Spef1; central microtubules; motile cilia.
© The Author(s) 2025. Published by Oxford University Press on behalf of China Science Publishing & Media Ltd.
Figures


References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
