A telomere-associated molecular landscape reveals immunological, microbial, and therapeutic heterogeneity in colorectal cancer
- PMID: 40492114
- PMCID: PMC12146184
- DOI: 10.3389/fmolb.2025.1615533
A telomere-associated molecular landscape reveals immunological, microbial, and therapeutic heterogeneity in colorectal cancer
Abstract
Background: Colorectal cancer (CRC) ranks among the most prevalent malignancies of the gastrointestinal tract and remains a leading cause of cancer-related mortality worldwide. Although telomere biology has been increasingly implicated in immune modulation and tumor progression, its clinical significance in CRC remains poorly understood.
Methods: We developed a telomere score, termed TELscore, by integrating transcriptomic and intratumoral microbiome profiles from publicly available colorectal cancer (CRC) cohorts. To comprehensively characterize TELscore subgroups, we performed pathway enrichment analysis, tumor immune microenvironment (TIME) profiling, and microbiome niche assessment. Whole-slide histopathological images (WSIs) and immunohistochemical (IHC) staining were utilized to visualize immune features, including tertiary lymphoid structures (TLSs), across subgroups. Patients were stratified into high and low TELscore categories, and the predictive robustness was validated across multiple independent training and validation cohorts. Chemotherapeutic drug sensitivity was evaluated using pharmacogenomic data from the Genomics of Drug Sensitivity in Cancer (GDSC) database. Furthermore, the predictive capacity of TELscore for immunotherapy response was independently assessed in an external cohort. Finally, single-cell RNA sequencing (scRNA-seq) analysis was conducted to further dissect the cellular landscape and immunological heterogeneity within the TME.
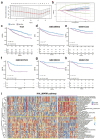
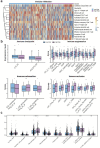
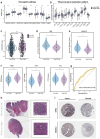
Results: TELscore stratified patients into two biologically and clinically distinct subgroups. The high TELscore group, which exhibited significantly shorter DFS, showed marked enrichment of tumorigenic pathways such as EMT, along with a distinctly immunosuppressive TME. This was reflected by elevated ESTIMATE/TIDE scores and corroborated by CIBERSORT, which revealed increased infiltration of M0 macrophages and upregulation of immunosuppressive signatures. In contrast, the low TELscore group was enriched for cell cycle related pathways, including E2F targets and the G2/M checkpoint, and demonstrated higher infiltration of pro-inflammatory M1 macrophages. 16S rRNA sequencing further revealed a divergent intratumoral microbiome between subgroups, the high TELscore group harbored significantly greater relative abundance of Selenomonas and Lachnoclostridium, two pathogenic genera previously associated with colorectal tumorigenesis. Complementary histopathological assessment via WSI demonstrated a marked absence of intraTLSs in high TELscore tumors. From a therapeutic standpoint, high TELscore tumors exhibited reduced sensitivity to standard chemotherapeutic agents-including Fluorouracil, Irinotecan, Oxaliplatin, and Docetaxel-as reflected by elevated IC50 values. Conversely, these tumors demonstrated increased susceptibility to MAPK pathway inhibitors, such as Selumetinib and Trametinib. Notably, TELscore also served as a robust predictor of immunotherapy response, which was validated in the IMvigor210 cohort. Finally, scRNA analysis highlighted profound cellular and functional divergence between TELscore subgroups. We identified intensified intercellular communication between inflammatory macrophages and fibroblasts, reinforcing the presence of an immunosuppressive niche.
Conclusion: TELscore is a robust stratification tool that captures the interplay between tumor biology, immune characteristics, and microbial ecology in colorectal cancer. By identifying clinically relevant subtypes with distinct therapeutic vulnerabilities, TELscore offers a powerful framework to advance personalized treatment and precision oncology.
Keywords: colorectal cancer; microbiome; prognosis; telomere; tumor microenvironment.
Copyright © 2025 Zhang, Fan, Zhao, Zhu, Xia and Xu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
A glutamine metabolism gene signature with prognostic and predictive value for colorectal cancer survival and immunotherapy response.Front Mol Biosci. 2025 May 15;12:1599141. doi: 10.3389/fmolb.2025.1599141. eCollection 2025. Front Mol Biosci. 2025. PMID: 40443528 Free PMC article.
-
Spatial and single-cell transcriptomic analysis reveals fibroblasts dependent immune environment in colorectal cancer.Biofactors. 2025 Mar-Apr;51(2):e70012. doi: 10.1002/biof.70012. Biofactors. 2025. PMID: 40068177
-
Identification of intratumoral microbiome-driven immune modulation and therapeutic implications in diffuse large B-cell lymphoma.Cancer Immunol Immunother. 2025 Mar 3;74(4):131. doi: 10.1007/s00262-025-03972-x. Cancer Immunol Immunother. 2025. PMID: 40029433 Free PMC article.
-
Transcriptomic correlates of cell cycle checkpoints with distinct prognosis, molecular characteristics, immunological regulation, and therapeutic response in colorectal adenocarcinoma.Front Immunol. 2023 Dec 8;14:1291859. doi: 10.3389/fimmu.2023.1291859. eCollection 2023. Front Immunol. 2023. PMID: 38143740 Free PMC article.
-
Deciphering colorectal cancer immune microenvironment transcriptional landscape on single cell resolution - A role for immunotherapy.Front Immunol. 2022 Aug 10;13:959705. doi: 10.3389/fimmu.2022.959705. eCollection 2022. Front Immunol. 2022. PMID: 36032085 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

