This is a preprint.
A genome-wide in vivo CRISPR screen identifies neuroprotective strategies in the mouse and human retina
- PMID: 40492201
- PMCID: PMC12148056
- DOI: 10.1101/2025.03.22.644712
A genome-wide in vivo CRISPR screen identifies neuroprotective strategies in the mouse and human retina
Abstract
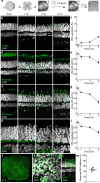
Retinitis pigmentosa (RP) is a genetically diverse blinding disorder lacking broadly effective therapies. We performed a genome-wide in vivo CRISPR knockout screen in mice carrying the P23H rhodopsin mutation (the most common cause of autosomal dominant RP in the United States) to systematically identify neuroprotective genes. We discovered multiple knockouts that accelerated rod photoreceptor loss, validated top candidates, and showed that overexpressing two genes-UFD1 and UXT-preserved rods and cones, maintained retinal function, and improved visual behaviors. To accelerate translation, we developed a human P23H RP model in adult retinal explants, recreating key disease features. UFD1 and UXT augmentation prevented photoreceptor loss in human P23H retinas. Our findings establish a pipeline for systematic identification and translational testing of neuroprotective genes in mouse and human RP models, provide a novel set of validated candidate genes, and underscore the therapeutic promise of UFD1 and UXT as mutation-agnostic strategies to preserve vision.
Figures










References
-
- Chaumet-Riffaud A.E., et al. Impact of Retinitis Pigmentosa on Quality of Life, Mental Health, and Employment Among Young Adults. Am J Ophthalmol 177, 169–174 (2017). - PubMed
-
- Daiger S.P., Sullivan L.S., Bowne S.J. & Rossiter B.J.F. RetNet: retinal information network. (2022).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
