Phage steering in the presence of a competing bacterial pathogen
- PMID: 40492711
- PMCID: PMC12211013
- DOI: 10.1128/spectrum.02882-24
Phage steering in the presence of a competing bacterial pathogen
Abstract
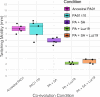
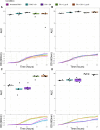
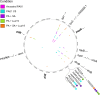
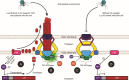
The rise of antibiotic-resistant bacteria has necessitated the development of alternative therapeutic strategies, such as bacteriophage therapy, where viruses infect bacteria, reducing bacterial burden. However, rapid bacterial resistance to phage treatment remains a critical challenge, potentially leading to failure. Phage steering, which leverages the evolutionary dynamics between phage and bacteria, offers a novel solution by driving bacteria to evolve away from virulence factors or resistance mechanisms. In this study, we examined whether phage steering using bacteriophage Luz19 could function in the presence of a competing pathogen, Staphylococcus aureus (SA) (USA300), while targeting Pseudomonas aeruginosa (PAO1). Through in vitro co-evolution experiments with and without the competitor, we observed that Luz19 consistently steered P. aeruginosa away from the Type IV pilus (T4P), a key virulence factor, without interference from SA. Genomic analyses revealed mutations in T4P-associated genes, including pilR and pilZ, which conferred phage resistance. Our findings suggest that phage steering remains effective even in polymicrobial environments, providing a promising avenue for enhancing bacteriophage therapy efficacy in complex infections.IMPORTANCEPhage steering-using phages that bind essential virulence or resistance-associated structures-offers a promising solution by selecting for resistance mutations that attenuate pathogenic traits. However, it remains unclear whether this strategy remains effective in polymicrobial contexts, where interspecies interactions may alter selective pressures. Here, we demonstrate that Pseudomonas aeruginosa evolves phage resistance via loss-of-function mutations in Type IV pilus (T4P) when challenged with the T4P-binding phage Luz19 and that this evolutionary trajectory is preserved even in the presence of a competing pathogen, Staphylococcus aureus. Phage resistance was phenotypically confirmed via twitching motility assays and genotypically via whole-genome sequencing. These findings support the robustness of phage steering under interspecies competition, underscoring its translational potential for managing complex infections-such as those seen in cystic fibrosis-where microbial diversity is the norm.
Keywords: bacteriophage evolution; bacteriophage therapy; bacteriophages; steering; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Bacteriophage infection drives loss of β-lactam resistance in methicillin-resistant Staphylococcus aureus.Elife. 2025 Jul 10;13:RP102743. doi: 10.7554/eLife.102743. Elife. 2025. PMID: 40637714 Free PMC article.
-
Lytic bacteriophages targeting multidrug-resistant Pseudomonas aeruginosa in Moschus berezovskii: isolation, characterization, and therapeutic efficacy against bacteremia.Virol J. 2025 Aug 19;22(1):285. doi: 10.1186/s12985-025-02715-9. Virol J. 2025. PMID: 40830499 Free PMC article.
-
Metapopulation model of phage therapy of an acute Pseudomonas aeruginosa lung infection.mSystems. 2024 Oct 22;9(10):e0017124. doi: 10.1128/msystems.00171-24. Epub 2024 Sep 4. mSystems. 2024. PMID: 39230264 Free PMC article.
-
Antibiotic strategies for eradicating Pseudomonas aeruginosa in people with cystic fibrosis.Cochrane Database Syst Rev. 2017 Apr 25;4(4):CD004197. doi: 10.1002/14651858.CD004197.pub5. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2023 Jun 2;6:CD004197. doi: 10.1002/14651858.CD004197.pub6. PMID: 28440853 Free PMC article. Updated.
-
Phage-Antibiotic Combinations for Pseudomonas: Successes in the Clinic and In Vitro Tenuously Connected.Microb Biotechnol. 2025 Jul;18(7):e70193. doi: 10.1111/1751-7915.70193. Microb Biotechnol. 2025. PMID: 40635382 Free PMC article. Review.
References
-
- Rohde C, Resch G, Pirnay J-P, Blasdel BG, Debarbieux L, Gelman D, Górski A, Hazan R, Huys I, Kakabadze E, Łobocka M, Maestri A, Almeida GM de F, Makalatia K, Malik DJ, Mašlaňová I, Merabishvili M, Pantucek R, Rose T, Štveráková D, Van Raemdonck H, Verbeken G, Chanishvili N. 2018. Expert opinion on three phage therapy related topics: bacterial phage resistance, phage training and prophages in bacterial production strains. Viruses 10:178. doi: 10.3390/v10040178 - DOI - PMC - PubMed
-
- Medel R, Mendez MA, Ossa CG, Botto-Mahan C. 2010. Arms race coevolution: the local and geographical structure of a host–parasite interaction. Evo Edu Outreach 3:26–31. doi: 10.1007/s12052-009-0191-7 - DOI
-
- Thompson JN. 2019. The geographic mosaic of coevolution . University of Chicago Press.
-
- Thompson JN, Pagel M. 2002. Coevolution. In Encyclopedia of evolution. Oxford University Press, Oxford.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

