Host-pathogen interaction profiling of nontypeable Haemophilus influenzae and Moraxella catarrhalis coinfection of bronchial epithelial cells
- PMID: 40492732
- PMCID: PMC12306180
- DOI: 10.1128/msphere.00242-25
Host-pathogen interaction profiling of nontypeable Haemophilus influenzae and Moraxella catarrhalis coinfection of bronchial epithelial cells
Abstract
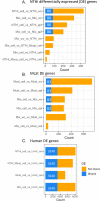
Chronic obstructive pulmonary disease (COPD) is a progressive inflammatory lung disease and the third leading cause of death globally. Nontypeable Haemophilus influenzae (NTHi) and Moraxella catarrhalis (Mcat) are common pathogens in individuals with COPD. Acquisition of NTHi or Mcat can cause acute exacerbations of COPD. NTHi and/or Mcat also persist for months in the lower airways and lead to chronic inflammation. We hypothesized that infection by NTHi or Mcat, singly or during coinfection, requires regulation of specific bacterial and host cell pathways. We investigated this phenomenon using an in vitro cell culture model consisting of lung carcinoma H292 cell lines, infected with NTHi, Mcat, or both species. Samples were fractionated into "apical fluid," containing free-floating bacteria, and adhered/invaded bacteria on or within H292 cells. We used transcriptomic profiling with RNA-seq and various bioinformatic analyses to disentangle host-pathogen interactions in epithelial cell infection from the perspective of each species. Several biological pathways were differentially regulated across all conditions (31, NTHi; 22, Mcat; and 169, human). NTHi transcriptomic profiles differed during mono-infection and coinfection; examples included downregulation of iron-sulfur metabolism (IscR regulon) and differential regulation of quorum sensing in coinfection compared to mono-infection. Mcat was comparatively less affected by the presence of NTHi during coinfection. H292 epithelial cells responded broadly to all infections with distinct responses to mono-infection and coinfection. Enriched host pathways included influenza/interferon/Wnt and proinflammatory responses. These findings suggest common and distinct processes involved in NTHi and/or Mcat-induced COPD pathogenesis and have implications for therapeutic intervention.IMPORTANCEChronic obstructive pulmonary disease (COPD) is a leading cause of death worldwide. Bacteria such as nontypeable Haemophilus influenzae (NTHi) and Moraxella catarrhalis (Mcat) can cause exacerbations of COPD, and they can persist in the lungs for months, which increases inflammation. We studied how these bacteria interact with lung cells by infecting a cell culture model with NTHi, Mcat, or both. We used RNA sequencing and bioinformatic analysis to examine how the bacteria and host cells respond. When NTHi and Mcat were present together, they behaved differently than when each was alone. We found that different host biological pathways were activated during infection, including those related to inflammation and immune responses. These results provide insights into how NTHi and Mcat contribute to COPD progression and suggest potential targets for new treatments.
Keywords: Haemophilus influenzae; Moraxella catarrhalis; RNA sequencing; chronic obstructive pulmonary disease; coinfection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Haemophilus influenzae oral vaccination for preventing acute exacerbations of chronic bronchitis and chronic obstructive pulmonary disease.Cochrane Database Syst Rev. 2014 Sep 9;(9):CD010010. doi: 10.1002/14651858.CD010010.pub2. Cochrane Database Syst Rev. 2014. Update in: Cochrane Database Syst Rev. 2017 Jun 19;6:CD010010. doi: 10.1002/14651858.CD010010.pub3. PMID: 25201571 Updated.
-
Haemophilus influenzae oral vaccination for preventing acute exacerbations of chronic bronchitis and chronic obstructive pulmonary disease.Cochrane Database Syst Rev. 2017 Jun 19;6(6):CD010010. doi: 10.1002/14651858.CD010010.pub3. Cochrane Database Syst Rev. 2017. PMID: 28626902 Free PMC article.
-
Modeling airway persistent infection of Moraxella catarrhalis and nontypeable Haemophilus influenzae by using human in vitro models.Front Cell Infect Microbiol. 2024 May 1;14:1397940. doi: 10.3389/fcimb.2024.1397940. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38751999 Free PMC article.
-
Comparative genomic analysis of emerging non-typeable Haemophilus influenzae (NTHi) causing emerging septic arthritis in Atlanta.PeerJ. 2025 Mar 21;13:e19081. doi: 10.7717/peerj.19081. eCollection 2025. PeerJ. 2025. PMID: 40130174 Free PMC article.
-
Inhaled steroids and risk of pneumonia for chronic obstructive pulmonary disease.Cochrane Database Syst Rev. 2014 Mar 10;2014(3):CD010115. doi: 10.1002/14651858.CD010115.pub2. Cochrane Database Syst Rev. 2014. PMID: 24615270 Free PMC article.
References
-
- Adeloye D, Song P, Zhu Y, Campbell H, Sheikh A, Rudan I, NIHR RESPIRE Global Respiratory Health Unit . 2022. Global, regional, and national prevalence of, and risk factors for, chronic obstructive pulmonary disease (COPD) in 2019: a systematic review and modelling analysis. Lancet Respir Med 10:447–458. doi: 10.1016/S2213-2600(21)00511-7 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

