Isolation and characterization of mollicute symbionts from a fungus-growing ant reveals high niche overlap leading to co-exclusion
- PMID: 40492740
- PMCID: PMC12239587
- DOI: 10.1128/mbio.00893-25
Isolation and characterization of mollicute symbionts from a fungus-growing ant reveals high niche overlap leading to co-exclusion
Abstract
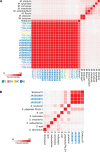
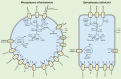
Two mollicute species belonging to the Mesoplasma and Spiroplasma genera have been detected in several species of fungus-growing ants using molecular methods. However, their ecological roles remain largely inferred from metagenomic data. To better understand their diversity and specialization, we cultured both of these Mesoplasma and Spiroplasma symbionts from the fungus-growing ant Trachymyrmex septentrionalis, providing the first isolated mollicutes from any fungus-growing ant species. The genomes of our isolates and related metagenome-assembled genomes (MAGs) from T. septentrionalis fungus gardens comprise two unique phylogenetic lineages compared to previously described Mesoplasma and Spiroplasma species, and from related MAGs previously sequenced from the leaf-cutting ant Acromyrmex echinatior. This suggests that the T. septentrionalis symbionts comprise undescribed species that can exclude each other from a niche that is largely shared between them. Mesoplasma genomes and MAGs also demonstrate regional specificity with their T. septentrionalis ant hosts. Both Mesoplasma and Spiroplasma strains from T. septentrionalis can catabolize glucose and fructose; both sugars are common in the ant's diet. Similarly, both these Mesoplasma and Spiroplasma can catabolize arginine, but only Mesoplasma can catabolize N-acetylglucosamine; both could produce ammonia for the ants or fungus garden. Based on our genomic and phenotypic analyses, we describe these T. septentrionalis symbionts as Mesoplasma whartonense sp. nov. and Spiroplasma attinicola sp. nov., providing insight into their genomic and phenotypic diversity and cultures to facilitate future studies of how these common but poorly understood members of the fungus-growing ant symbiosis separately colonize different ant colonies despite having highly overlapping niches.
Importance: Fungus-growing ants partner with multiple microbial symbionts to obtain food and remain free from disease. Of these symbionts, those inhabiting the ant gut remain the least understood and are known only from environmental surveys. Such surveys can infer potential functions of gut symbionts, but cultures are required to experimentally validate these hypotheses. Here, we describe the first cultures of the ant gut symbionts of the fungus-growing ant Trachymyrmex septentrionalis, using comparative genomics and phenotypic experiments to describe them as two novel species: Mesoplasma whartonense sp. nov. and Spiroplasma attinicola sp. nov. This genomic analysis suggests that these species are highly specialized to T. septentrionalis and are distinct from related environmental data generated from the related ant species Acromyrmex echinatior, implying substantial host specificity. Our phenotypic experiments and genomic reconstructions highlight the highly overlapping niches and likely costs and benefits of these symbionts to their ant host, setting the stage for further experimentation.
Keywords: Mesoplasma; Spiroplasma; comparative genomics; fungus-growing ants; host specialization; symbiosis; taxonomy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Douglas AE, Prosser WA. 1992. Synthesis of the essential amino acid tryptophan in the pea aphid (Acyrthosiphon pisum) symbiosis. J Insect Physiol 38:565–568. doi: 10.1016/0022-1910(92)90107-O - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
