Targeted next-generation sequencing: a promising approach for Mycobacterium tuberculosis detection and drug resistance when applied in paucibacillary clinical samples
- PMID: 40492765
- PMCID: PMC12211088
- DOI: 10.1128/spectrum.03127-24
Targeted next-generation sequencing: a promising approach for Mycobacterium tuberculosis detection and drug resistance when applied in paucibacillary clinical samples
Abstract
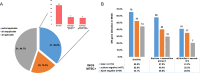
Tuberculosis (TB) returns to be the leading infectious killer globally after coronavirus disease 2019. The World Health Organization (WHO) formally included targeted next-generation sequencing (tNGS) in its list of recommendations for Mycobacterium tuberculosis (MTB) and drug resistance (DR). In this study, we explored the application of various clinical sample types for TB diagnosis and DR profiles. In comparison to the composite reference standard, the overall sensitivity values of culture, Xpert, metagenomic next-generation sequencing (mNGS), and tNGS were 0.458, 0.614, 0.772, and 0.760, respectively. tNGS had sensitivity similar to mNGS, which had advantages over culture and Xpert, respectively, despite different classification of sample types. In comparison to the microbiological reference standard, the overall sensitivity values of culture, Xpert, mNGS, and tNGS were 0.606, 0.811, 0.856, and 0.884, respectively. Suprisingly, in extrapulmonary tissue and serous effusion, mNGS and tNGS had advantages over Xpert. DR-related mutations were detected in 15 cases (13.2%). There were 51 (44.7%) applicable for all DR genes, with 22 (19.3%) not applicable for DR genes. DR genes were partially applicable in 41 (36.0%) samples. However, in culture-negative TB cases, tNGS can additionally provide 52.7% first-line DR profiles. Sanger sequencing was performed on 14 samples to confirm gene mutation identified by tNGS, and the results were entirely consistent. It was concluded that the tNGS assay was a promising approach in the initial diagnostic test of MTB and DR-related genes in different clinical samples, even for the smear- and culture-negative paucibacillary samples.IMPORTANCEtNGS combines gene-specific amplification with next-generation sequencing to detect MTB and drug-resistant genes by amplifying numerous loci directly from clinical samples. The WHO implemented tNGS for the purpose of monitoring respiratory specimens for MTB detection and DR-TB due to its high sensitivity and specificity, culture independence, and ability to report heterogeneous/silent mutations. The sensitivity outperformed both culture and Xpert, and the turnaround time was significantly less than that of culture-based assays. The tNGS assay used in this study costs USD 96 and has a 12 hour turnaround time. Nonetheless, tNGS has a great deal of promise for enhancing TB detection while also addressing DR strains.
Keywords: Mycobacterium tuberculosis; paucibacillary tuberculosis; targeted next-generation sequencing; tuberculosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Xpert® MTB/RIF assay for extrapulmonary tuberculosis and rifampicin resistance.Cochrane Database Syst Rev. 2018 Aug 27;8(8):CD012768. doi: 10.1002/14651858.CD012768.pub2. Cochrane Database Syst Rev. 2018. Update in: Cochrane Database Syst Rev. 2021 Jan 15;1:CD012768. doi: 10.1002/14651858.CD012768.pub3. PMID: 30148542 Free PMC article. Updated.
-
A comprehensive evaluation of a novel targeted-sequencing workflow for Mycobacterium species identification and anti-tuberculosis drug-resistance detection.Front Cell Infect Microbiol. 2025 Jun 9;15:1584237. doi: 10.3389/fcimb.2025.1584237. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40552123 Free PMC article.
-
Evaluating culture-free targeted next-generation sequencing for diagnosing drug-resistant tuberculosis: a multicentre clinical study of two end-to-end commercial workflows.Lancet Infect Dis. 2025 Mar;25(3):325-334. doi: 10.1016/S1473-3099(24)00586-3. Epub 2024 Oct 29. Lancet Infect Dis. 2025. PMID: 39486428
-
Xpert MTB/RIF Ultra assay for tuberculosis disease and rifampicin resistance in children.Cochrane Database Syst Rev. 2022 Sep 6;9(9):CD013359. doi: 10.1002/14651858.CD013359.pub3. Cochrane Database Syst Rev. 2022. PMID: 36065889 Free PMC article.
-
Xpert MTB/XDR for detection of pulmonary tuberculosis and resistance to isoniazid, fluoroquinolones, ethionamide, and amikacin.Cochrane Database Syst Rev. 2022 May 18;5(5):CD014841. doi: 10.1002/14651858.CD014841.pub2. Cochrane Database Syst Rev. 2022. PMID: 35583175 Free PMC article.
References
-
- WHO . 2024. Global tuberculosis report 2024
-
- Schön T, Matuschek E, Mohamed S, Utukuri M, Heysell S, Alffenaar J-W, Shin S, Martinez E, Sintchenko V, Maurer FP, Keller PM, Kahlmeter G, Köser CU. 2019. Standards for MIC testing that apply to the majority of bacterial pathogens should also be enforced for Mycobacterium tuberculosis complex. Clin Microbiol Infect 25:403–405. doi: 10.1016/j.cmi.2019.01.019 - DOI - PMC - PubMed
-
- WHO . 2014. Xpert MTB/RIF implementation manual: technical and operational ‘how-to’; practical considerations. - PubMed
-
- Miotto P, Tessema B, Tagliani E, Chindelevitch L, Starks AM, Emerson C, Hanna D, Kim PS, Liwski R, Zignol M, et al. 2017. A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis. Eur Respir J 50:1701354. doi: 10.1183/13993003.01354-2017 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

