Expression of full-length dystrophin reverses muscular dystrophy defects in young and old mdx4cv mice
- PMID: 40493400
- PMCID: PMC12321383
- DOI: 10.1172/JCI189075
Expression of full-length dystrophin reverses muscular dystrophy defects in young and old mdx4cv mice
Abstract
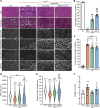
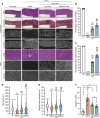
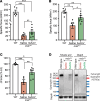
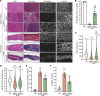
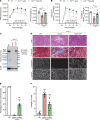
Gene replacement therapies mediated by adeno-associated viral (AAV) vectors represent a promising approach for treating genetic diseases. However, their modest packaging capacity (~4.7 kb) remains an important constraint and significantly limits their application for genetic disorders involving large genes. A prominent example is Duchenne muscular dystrophy (DMD), whose protein product dystrophin is generated from a 11.2 kb segment of the DMD mRNA. Here, we explored methods that enable efficient expression of full-length dystrophin via triple AAV codelivery. This method exploits the protein trans-splicing mechanism mediated by split inteins. We identified a combination of efficient and specific split intein pairs that enabled the reconstitution of full-length dystrophin from 3 dystrophin fragments. We show that systemic delivery of low doses of the myotropic AAVMYO1 in mdx4cv mice led to efficient expression of full-length dystrophin in the hind limb, diaphragm, and heart muscles. Notably, muscle morphology and physiology were significantly improved in triple-AAV-treated mdx4cv mice versus saline-treated controls. This method shows the feasibility of expressing large proteins from several fragments that were delivered using low doses of myotropic AAV vectors. It can be adapted to other large genes involved in disorders for which gene replacement remains challenged by the modest AAV cargo capacity.
Keywords: Gene therapy; Genetic diseases; Genetics; Muscle biology; Skeletal muscle; Therapeutics.
Conflict of interest statement
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

