Emerin is an effector of oncogenic KRAS-driven nuclear dynamics in pancreatic cancer
- PMID: 40493403
- PMCID: PMC12288966
- DOI: 10.1172/jci.insight.187799
Emerin is an effector of oncogenic KRAS-driven nuclear dynamics in pancreatic cancer
Abstract
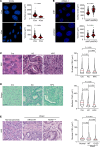
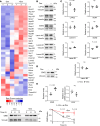
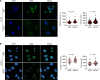
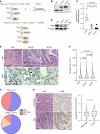
For over a century, scientists reported the disruption of normal nuclear shape and size in cancer. These changes have long been used as tools for diagnosis and staging of malignancies. However, to date, the mechanisms underlying these aberrant nuclear phenotypes and their biological significance remain poorly understood. Using a model of pancreatic ductal adenocarcinoma (PDAC), the major histological subtypes of pancreatic cancer, we found that oncogenic mutant KRAS reduces nuclear size. Transcriptomic and protein expression analysis of mutant KRAS-expressing PDAC cells revealed differential levels of several nuclear envelope-associated genes. Further analysis demonstrated the nuclear lamina protein, Emerin (EMD), acted downstream of KRAS to mediate nuclear size reduction in PDAC. Analysis of human PDAC samples showed that increased EMD expression associates with reduced nuclear size. Finally, in vivo genetic depletion of EMD in a mutant KRAS-driven PDAC model resulted in increased nuclear size and a reduced incidence of poorly differentiated PDAC. Thus, our data provide evidence of a potentially novel mechanism underlying nuclear size regulation and its effect in PDAC carcinogenesis.
Keywords: Cancer; Cell biology; Oncology.
Conflict of interest statement
Figures
References
-
- Schirmer EC, de las Heras JI, eds. Cancer Biology and the Nuclear Envelope. Springer; 2014.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

