Wilms tumor 1 impairs apoptotic clearance of fibroblasts in distal fibrotic lung lesions
- PMID: 40493404
- PMCID: PMC12321392
- DOI: 10.1172/JCI188819
Wilms tumor 1 impairs apoptotic clearance of fibroblasts in distal fibrotic lung lesions
Abstract
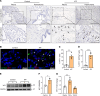
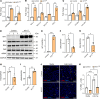
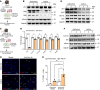
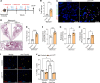
Idiopathic pulmonary fibrosis (IPF) is a fatal fibrotic lung disease characterized by impaired fibroblast clearance and excessive extracellular matrix (ECM) protein production. Wilms tumor 1 (WT1), a transcription factor, is selectively upregulated in IPF fibroblasts. However, the mechanisms by which WT1 contributes to fibroblast accumulation and ECM production remain unknown. Here, we investigated the heterogeneity of WT1-expressing mesenchymal cells using single-nucleus RNA-Seq of distal lung tissues from patients with IPF and control donors. WT1 was selectively upregulated in a subset of IPF fibroblasts that coexpressed several prosurvival and ECM genes. The results of both loss-of-function and gain-of-function studies were consistent with a role for WT1 as a positive regulator of prosurvival genes to impair apoptotic clearance and promote ECM production. Fibroblast-specific overexpression of WT1 augmented fibroproliferation, myofibroblast accumulation, and ECM production during bleomycin-induced pulmonary fibrosis in young and aged mice. Together, these findings suggest that targeting WT1 is a promising strategy for attenuating fibroblast expansion and ECM production during fibrogenesis.
Keywords: Apoptosis; Extracellular matrix; Fibrosis; Inflammation; Pulmonology.
Conflict of interest statement
Figures




Update of
-
Wilms' tumor 1 impairs apoptotic clearance of fibroblasts in distal fibrotic lung lesions.bioRxiv [Preprint]. 2025 May 22:2025.05.17.654673. doi: 10.1101/2025.05.17.654673. bioRxiv. 2025. Update in: J Clin Invest. 2025 Jun 10;135(15):e188819. doi: 10.1172/JCI188819. PMID: 40475429 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

