Cord blood proteomics identifies biomarkers of early-onset neonatal sepsis
- PMID: 40493423
- PMCID: PMC12288901
- DOI: 10.1172/jci.insight.193826
Cord blood proteomics identifies biomarkers of early-onset neonatal sepsis
Abstract
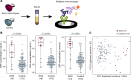
BACKGROUNDSymptoms of early-onset neonatal sepsis (EOS) in preterm infants are nonspecific and overlap with normal postnatal physiological adaptations and noninfectious pathologies. This clinical uncertainty and the lack of reliable EOS diagnostics results in liberal use of antibiotics in the first days to weeks of life, leading to increased risk of antibiotic-related morbidities in infants who do not have an invasive infection. METHODSTo identify potential biomarkers for EOS in newborn infants, we used unlabeled tandem mass spectrometry proteomics to identify differentially abundant proteins in the umbilical cord blood of infants with and without culture-confirmed EOS. Proteins were then confirmed using immunoassay, and logistic regression and random forest models were built, including both biomarker concentration and clinical variables to predict EOS. RESULTSThese data identified 5 proteins that were significantly upregulated in infants with EOS, 3 of which (serum amyloid A, C-reactive protein, and lipopolysaccharide-binding protein) were confirmed using a quantitative immunoassay. The random forest classifier for EOS was applied to a cohort of infants with culture-negative presumed sepsis. Most infants with presumed sepsis were classified as resembling infants in the control group, with low EOS biomarker concentrations.CONCLUSIONThese results suggest that cord blood biomarker screening may be useful for early stratification of EOS risk among neonates, improving targeted, evidence-based use of antibiotics early in life. FUNDINGNIH, Gerber Foundation, Friends of Prentice, Thrasher Research Fund, Ann & Robert H. Lurie Children's Hospital, and Stanley Manne Children's Research Institute of Lurie Children's.
Keywords: Bacterial infections; Biomarkers; Immunology; Infectious disease; Proteomics.
Conflict of interest statement
Figures




References
-
- WHO. Global report on the epidemiology and burden of sepsis: current evidence, identifying gaps and future directions. https://www.who.int/publications/i/item/9789240010789 Published September 9, 2020. Accessed June 5, 2025.
MeSH terms
Substances
Grants and funding
- R01 AI165236/AI/NIAID NIH HHS/United States
- R01 AI150455/AI/NIAID NIH HHS/United States
- R01 HL139798/HL/NHLBI NIH HHS/United States
- K23 HL093302/HL/NHLBI NIH HHS/United States
- R21 AI163912/AI/NIAID NIH HHS/United States
- P41 GM108569/GM/NIGMS NIH HHS/United States
- U19 AI135964/AI/NIAID NIH HHS/United States
- UL1 TR001422/TR/NCATS NIH HHS/United States
- P30 CA060553/CA/NCI NIH HHS/United States
- R01 AI150998/AI/NIAID NIH HHS/United States
- R01 AI177498/AI/NIAID NIH HHS/United States
- K23 AI139337/AI/NIAID NIH HHS/United States
- S10 OD025194/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials

