Fine-scale population structure of Japanese Helicobacter pylori provides new anthropological and epidemiological insights
- PMID: 40493491
- PMCID: PMC12152251
- DOI: 10.1099/mgen.0.001419
Fine-scale population structure of Japanese Helicobacter pylori provides new anthropological and epidemiological insights
Abstract
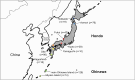
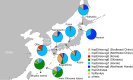
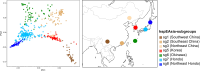
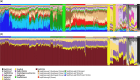
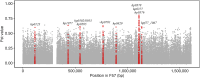
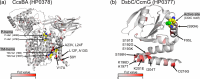
Helicobacter pylori is considered to contribute to gastric cancer and is also used as a marker to trace human migration due to its co-evolution with humans. To understand the recently proposed tripartite model suggesting three ancestral origins for the Japanese population and address the enigma of the high incidence of gastric cancer in Northeast Hondo (Hondo is mainland Japan), we conducted a fine-scale population structure analysis using a large Japanese H. pylori dataset, including 438 strains from 9 regions based on whole-genome sequences. As a result of fineSTRUCTURE analysis, it was found that H. pylori in Northeast Hondo is genetically distinct from hspEAsia subgroup 7 (sg7), which is widely distributed elsewhere in Hondo. We named this new subgroup hspEAsia-sg8 (Northeast Hondo). Ancestry analysis using ChromoPainter revealed that, while a large proportion of the genomes of hspEAsia-sg8 strains were painted by donors from their own population, the ancestry components of hspEAsia-sg7 showed a high proportion of Chinese and Korean components, suggesting that they were formed through admixture with continental hspEAsia subgroups. These results align with human genome studies, which indicate an original ancestry component in Northeast Hondo and a higher proportion of East Asian components in West Hondo, supporting the tripartite model. This also suggests novel potential for biogeographic ancestry inference in forensic science, as the H. pylori genome can distinguish Hondo populations. Furthermore, fixation index analysis comparing the genome of hspEAsia-sg8 with other Japanese hspEAsia subgroups revealed a high number of nonsynonymous mutations in hp0378 (ccsBA) and hp0377 (dsbC/ccmG). Because these genes are involved in cytochrome c maturation and disulphide bond formation, the detected mutations may affect bacterial survival, growth or pathogenicity. This study supports the tripartite model for the formation of modern Japanese people and suggests that the strain of H. pylori prevalent in the Northeast Hondo region may contribute to the high incidence of gastric cancer there.
Keywords: Helicobacter pylori; ancestry analysis; gastric cancer; human migration; population structure.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

References
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

