Extended-spectrum β-lactamase-producing Enterobacterales among people living with human immunodeficiency virus across the globe: A systematic review and meta-analysis
- PMID: 40493608
- PMCID: PMC12151346
- DOI: 10.1371/journal.pone.0321873
Extended-spectrum β-lactamase-producing Enterobacterales among people living with human immunodeficiency virus across the globe: A systematic review and meta-analysis
Abstract
Background: People living with HIV are vulnerable to antibiotic-resistant bacterial infections because of frequent healthcare visits, the consumption of many antimicrobials, and the weakened immune system to fight infections. Our objective was to provide comprehensive data about ESBL-producing Enterobacterales among HIV-positive individuals across the globe.
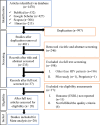
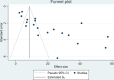
Methods: This meta-analysis was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. To select eligible articles published between January 1, 2010, and May 12, 2024, a literature search was performed on available electronic databases such as PubMed, Hinari, Google Scholar, and Scopus. The quality of the included studies was assessed via the Joanna Briggs Institute critical appraisal tool. The data were extracted from the eligible studies via Microsoft Excel 2019 and analyzed via STATA version 17. A random effects model was constructed via the DerSimonian and Laird method. The heterogeneity was checked through the Cochrane Q statistic, and the magnitude was quantitatively measured via I2 statistics. To determine the possible sources of heterogeneity, a subgroup analysis was performed. Additionally, a sensitivity analysis was conducted, and publication bias was checked via funnel plots and Egger's regression tests. A p value of less than 0.05 was considered evidence of heterogeneity and small study effects according to the Cochrane Q statistic and Egger's test, respectively. The protocol was registered previously (PROSPERO ID: CRD42024557981).
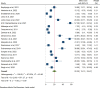
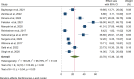
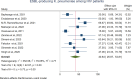
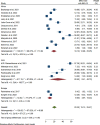
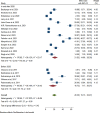
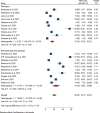
Results: A total of 5305 HIV-positive individuals from 20 studies were included in our meta-analysis. The overall pooled prevalence of ESBL-producing Enterobacterales among HIV-positive individuals was 20.30% (931/5305; 95% CI: 15.13-25.47%, P < 0.001), with a high level of heterogeneity (I2 = 97.82%, P < 0.001). The predominant ESBL producers were K. pneumoniae, with a pooled prevalence of 40.84% (76/217; 95% CI: 26.87-54.81%), followed closely by E. coli at 40.14% (348/985; 95% CI: 27.83-52.45%). In the subgroup analysis, the highest magnitude of ESBL-producing pathogens was observed in Asia (195/715; 28.55%), followed by Africa (666/3981; 19.12%). Additionally, the highest pooled prevalence of ESBL-producing pathogens among HIV-positive individuals was reported to be colonization 23.78% (613/2455; 95% CI: 15.36-32.19, I² = 96.78%, p < 0.001), followed by infection 15.77% (318/2850; 95% CI: 10.06-21.49, I² = 97.45%, p < 0.001). Among the different types of ESBL enzyme-encoding genes, blaCTX-M was the most common (73 out of 150 isolates, 48.7%), followed by blaTEM (49 out of 150, 32.7%).
Conclusion and recommendations: This study demonstrated that HIV-positive individuals are commonly colonized and infected with ESBL-producing Enterobacterales. The highest prevalence of these pathogens was reported in Asia and Africa. To reduce mortality from severe bacterial infections in HIV patients, resources should be distributed equitably across all regions. Particular attention should be given to high-prevalence areas, where early detection of colonization and infection with antibiotic-resistant pathogens is critical. Enhanced surveillance of ESBL-producing organisms among HIV-positive individuals is also strongly recommended.
Copyright: © 2025 Tigabie et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Colonization with extended-spectrum β-lactamase and carbapenemase-producing Enterobacterales in Ethiopia: A systematic review and meta-analysis.PLoS One. 2025 Apr 1;20(4):e0316492. doi: 10.1371/journal.pone.0316492. eCollection 2025. PLoS One. 2025. PMID: 40168361 Free PMC article.
-
Extended-spectrum beta-lactamase and carbapenemase-producing Gram-negative bacteria in urinary tract infections in Ethiopia: a systematic review and meta-analysis.BMC Urol. 2025 Jan 21;25(1):11. doi: 10.1186/s12894-025-01695-w. BMC Urol. 2025. PMID: 39838325 Free PMC article.
-
Prevalence and molecular characterization of ESBL-producing Enterobacteriaceae in Egypt: a systematic review and meta-analysis of hospital and community-acquired infections.Antimicrob Resist Infect Control. 2024 Dec 5;13(1):145. doi: 10.1186/s13756-024-01497-z. Antimicrob Resist Infect Control. 2024. PMID: 39639352 Free PMC article.
-
Folic acid supplementation and malaria susceptibility and severity among people taking antifolate antimalarial drugs in endemic areas.Cochrane Database Syst Rev. 2022 Feb 1;2(2022):CD014217. doi: 10.1002/14651858.CD014217. Cochrane Database Syst Rev. 2022. PMID: 36321557 Free PMC article.
-
Faecal carriage of extended-spectrum beta-lactamase and carbapenemase-producing enterobacterales among HIV patients at Jimma Medical Center, Southwest Ethiopia.BMC Microbiol. 2024 Nov 6;24(1):459. doi: 10.1186/s12866-024-03596-8. BMC Microbiol. 2024. PMID: 39506640 Free PMC article.
References
-
- World Health Organization. Ten threats to global health 2019. [Available from: https://www.who.int/news-room/spotlight/ten-threats-to-global-health-in-....
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

