Role of the SAF-A/HNRNPU SAP domain in X chromosome inactivation, nuclear dynamics, transcription, splicing, and cell proliferation
- PMID: 40493679
- PMCID: PMC12176297
- DOI: 10.1371/journal.pgen.1011719
Role of the SAF-A/HNRNPU SAP domain in X chromosome inactivation, nuclear dynamics, transcription, splicing, and cell proliferation
Abstract
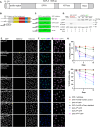
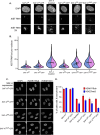
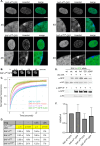
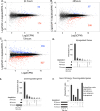
SAF-A/HNRNPU is conserved throughout vertebrates and has emerged as an important factor regulating a multitude of nuclear functions, including lncRNA localization, gene expression, and splicing. Here we show the SAF-A protein is highly dynamic and interacts with nascent transcripts as part of this dynamic movement. This finding revises current models of SAF-A: rather than being part of a static nuclear scaffold/matrix structure that acts as a stable tether between RNA and chromatin, SAF-A executes nuclear functions as a dynamic protein, suggesting contacts between SAF-A, RNA, and chromatin are more high turnover interactions than previously appreciated. SAF-A has several functional domains, including an N-terminal SAP domain that binds directly to DNA and RNA. Phosphorylation of SAP domain serines S14 and S26 is important for SAF-A localization and function during mitosis, however, whether these serines are involved in interphase functions of SAF-A is not known. In this study we tested for the role of the SAP domain, and SAP domain serines S14 and S26 in X chromosome inactivation, protein dynamics, gene expression, splicing, and cell proliferation. Here we show that the SAP domain, and SAP domain serines S14 and S26 are required to maintain XIST RNA localization and XIST-dependent histone modifications on the inactive X chromosome, to execute normal protein dynamics, and to maintain normal cell proliferation. In addition, we present evidence that a Xi localization signal resides in the SAP domain, enabling SAF-A to engage with the Xi compartment in a manner distinct from other nuclear territories. We found that the SAP domain is not required to maintain gene expression and plays only a minor role in mRNA splicing. We propose a model whereby dynamic phosphorylation of SAF-A serines S14 and S26 mediates rapid turnover of SAF-A interactions with nuclear structures during interphase. Our data suggest that different nuclear compartments may have distinct requirements for the SAF-A SAP domain to execute nuclear functions, a level of control that was not previously known.
Copyright: © 2025 Sharp et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Update of
-
Role of the SAF-A SAP domain in X inactivation, transcription, splicing, and cell proliferation.bioRxiv [Preprint]. 2024 Sep 10:2024.09.09.612041. doi: 10.1101/2024.09.09.612041. bioRxiv. 2024. Update in: PLoS Genet. 2025 Jun 10;21(6):e1011719. doi: 10.1371/journal.pgen.1011719. PMID: 39314300 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

