NF1 Loss Promotes EGFR Activation and Confers Sensitivity to EGFR Inhibition in NF1-Mutant Melanoma
- PMID: 40494652
- PMCID: PMC12221223
- DOI: 10.1158/0008-5472.CAN-24-3904
NF1 Loss Promotes EGFR Activation and Confers Sensitivity to EGFR Inhibition in NF1-Mutant Melanoma
Abstract
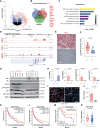
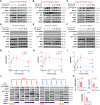
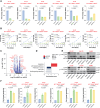
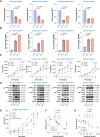
Targeted therapies and immunotherapy have improved treatment outcomes for many patients with melanoma. However, patients whose melanomas harbor driver mutations in the neurofibromin 1 (NF1) tumor suppressor gene often lack effective targeted treatment options when their tumors do not respond to immunotherapy. In this study, we utilized patient-derived short-term cultures and multiomics approaches to identify molecular features that could inform the development of therapies for patients with NF1-mutant (NF1Mut) melanoma. Differential gene expression analysis revealed that EGFR is highly expressed and active in NF1Mut melanoma cells, in which it hyperactivates ERK and AKT, leading to increased tumor cell proliferation, survival, and growth. In contrast, genetic or pharmacologic inhibition of EGFR hindered cell proliferation and survival and suppressed tumor growth in patient-derived NF1Mut melanoma models but not in NF1 wild-type models. These results reveal a connection between NF1 loss and increased EGFR expression that is critical for the survival and growth of NF1Mut melanoma cells in patient-derived culture and xenograft models, irrespective of their BRAF and NRAS mutation status.
Significance: NF1 mutant melanomas rely on EGFR activation and can be effectively treated with the EGFR inhibitors cetuximab or afatinib, supporting further testing in clinical trials.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
No disclosures were reported.
Figures



References
-
- Curti BD, Faries MB. Recent advances in the treatment of melanoma. N Engl J Med 2021;384:2229–40. - PubMed
-
- Luke JJ, Flaherty KT, Ribas A, Long GV. Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nat Rev Clin Oncol 2017;14:463–82. - PubMed
-
- Jour G, Illa-Bochaca I, Ibrahim M, Donnelly D, Zhu K, Miera EV, et al. Genomic and transcriptomic analyses of NF1-mutant melanoma identify potential targeted approach for treatment. J Invest Dermatol 2023;143:444–55.e8. - PubMed
-
- Dummer R, Ascierto PA, Gogas HJ, Arance A, Mandala M, Liszkay G, et al. Encorafenib plus binimetinib versus vemurafenib or encorafenib in patients with BRAF-mutant melanoma (COLUMBUS): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2018;19:603–15. - PubMed
MeSH terms
Substances
Grants and funding
- R01 CA274100/CA/NCI NIH HHS/United States
- P30 CA016087/CA/NCI NIH HHS/United States
- R01 CA259295/CA/NCI NIH HHS/United States
- P50 CA225450/CA/NCI NIH HHS/United States
- U54 CA263001/CA/NCI NIH HHS/United States
- R01 CA266656/CA/NCI NIH HHS/United States
- NCI P50 CA225450/Center for Cancer Research (CCR)
- U54 CA2630001/Center for Cancer Research (CCR)
- Fellow Research Grants 1287389/Melanoma Research Foundation (MRF)
- R01 CA274100/CA/NCI NIH HHS/United States
- R01 CA259295/CA/NCI NIH HHS/United States
- R01 CA266656/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

