Seasonal transcriptome profile across three different tissues of the Andean Killifish Orestias ascotanensis
- PMID: 40494877
- PMCID: PMC12152186
- DOI: 10.1038/s41597-025-05345-6
Seasonal transcriptome profile across three different tissues of the Andean Killifish Orestias ascotanensis
Abstract
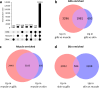
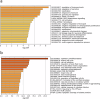
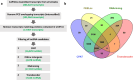
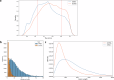
The Andean killifish Orestias ascotanensis inhabits the high-altitude Ascotán Salt Pan, an environment with variable salinity, high UV exposure, low oxygen, and extreme daily temperature fluctuations. These conditions make it an excellent model for studying high-altitude fish biology. However, the transcriptomic responses of O. ascotanensis to seasonal acclimation remain unexplored. To investigate seasonal and tissue-specific transcriptomic profiles, RNA-seq was performed on 42 libraries from gills, skin, and muscle tissues of 14 individuals collected in summer (n = 7) and winter (n = 7). Each library had a median of 105 million reads. Principal component analysis revealed strong tissue-specific expression, and seasonal differential expression analyses identified significant transcriptomic changes within each tissue. Additionally, a bioinformatics pipeline identified 10,365 high-confidence long non-coding RNAs (lncRNAs), predicted by at least three computational tools. Compared to protein-coding genes, lncRNAs exhibited higher tissue specificity, with a predominance of monoexonic structures and shorter exon lengths. This dataset provides the first comprehensive view of seasonal mRNA and lncRNA expression in O. ascotanensis tissues.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Cabrol, N. A. et al. The High-Lakes Project. J Geophys Res Biogeosci114 (2009).
-
- Collado, G. A. & Méndez, M. A. Microgeographic differentiation among closely related species of Biomphalaria (Gastropoda: Planorbidae) from the Andean Altiplano. Zool J Linn Soc10.1111/zoj.12073 (2013).
-
- Collado, G. A., Valladares, M. A. & Méndez, M. A. A new species of Heleobia (Caenogastropoda: Cochliopidae) from the Chilean Altiplano. Zootaxa Preprint at 10.11646/zootaxa.4137.2.8 (2016). - PubMed
-
- Sáez, P. A. et al. A new endemic lineage of the Andean frog genus Telmatobius (Anura, Telmatobiidae) from the western slopes of the central Andes. Zool J Linn Soc10.1111/zoj.12152 (2014).
-
- Parenti, L. A taxonomic revision of the Andean killifish genus Orestias (Cyprinodontiformes, Cyprinodontidae). Bull Am Mus Nat Hist (1984).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

