Retinoic acid breakdown is required for proximodistal positional identity during axolotl limb regeneration
- PMID: 40494878
- PMCID: PMC12152164
- DOI: 10.1038/s41467-025-59497-5
Retinoic acid breakdown is required for proximodistal positional identity during axolotl limb regeneration
Abstract
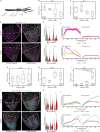
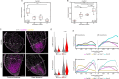
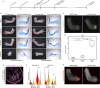
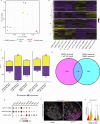
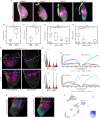
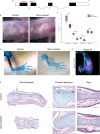
Regenerating limbs retain their proximodistal (PD) positional identity following amputation. This positional identity is genetically encoded by PD patterning genes that instruct blastema cells to regenerate the appropriate PD limb segment. Retinoic acid (RA) is known to specify proximal limb identity, but how RA signaling levels are established in the blastema is unknown. Here, we show that RA breakdown via CYP26B1 is essential for determining RA signaling levels within blastemas. CYP26B1 inhibition molecularly reprograms distal blastemas into a more proximal identity, phenocopying the effects of administering excess RA. We identify Shox as an RA-responsive gene that is differentially expressed between proximally and distally amputated limbs. Ablation of Shox results in shortened limbs with proximal skeletal elements that fail to initiate endochondral ossification. These results suggest that PD positional identity is determined by RA degradation and RA-responsive genes that regulate PD skeletal element formation during limb regeneration.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


Update of
-
Retinoic acid breakdown is required for proximodistal positional identity during amphibian limb regeneration.bioRxiv [Preprint]. 2024 Aug 9:2024.08.07.607055. doi: 10.1101/2024.08.07.607055. bioRxiv. 2024. Update in: Nat Commun. 2025 Jun 10;16(1):4798. doi: 10.1038/s41467-025-59497-5. PMID: 39149228 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- R01HD099174/U.S. Department of Health & Human Services | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- F31 HD116512/HD/NICHD NIH HHS/United States
- R01 HD099174/HD/NICHD NIH HHS/United States
- P40 OD019794/OD/NIH HHS/United States
- 1656429/National Science Foundation (NSF)
LinkOut - more resources
Full Text Sources

