Iterative transcription factor screening enables rapid generation of microglia-like cells from human iPSC
- PMID: 40494892
- PMCID: PMC12152180
- DOI: 10.1038/s41467-025-59596-3
Iterative transcription factor screening enables rapid generation of microglia-like cells from human iPSC
Abstract
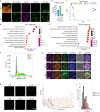
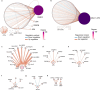
Differentiation of induced pluripotent stem cells (iPSCs) into specialized cell types is essential for uncovering cell-type specific molecular mechanisms and interrogating cellular function. Transcription factor screens have enabled efficient production of a few cell types; however, engineering cell types that require complex transcription factor combinations remains challenging. Here, we report an iterative, high-throughput single-cell transcription factor screening method that enables the identification of transcription factor combinations for specialized cell differentiation, which we validated by differentiating human microglia-like cells. We found that the expression of six transcription factors, SPI1, CEBPA, FLI1, MEF2C, CEBPB, and IRF8, is sufficient to differentiate human iPSC into cells with transcriptional and functional similarity to primary human microglia within 4 days. Through this screening method, we also describe a novel computational method allowing the exploration of single-cell RNA sequencing data derived from transcription factor perturbation assays to construct causal gene regulatory networks for future cell fate engineering.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: S.L. and G.M.C. are listed as inventors of a patent related to work on this article. G.M.C., P.K., and A.H.M.N. are co-founders/employees/advisors at, and have equity in GC Therapeutics, Inc, and are inventors on patents filed by the Presidents and Fellows of Harvard College. Full disclosure for GMC is available at arep.med.harvard.edu/gmc/tech.html . The remaining authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

