Ki-67 promotes inflammatory signaling governing neutrophil recruitment during respiratory infections
- PMID: 40494897
- PMCID: PMC12340004
- DOI: 10.1038/s44321-025-00261-z
Ki-67 promotes inflammatory signaling governing neutrophil recruitment during respiratory infections
Abstract
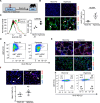
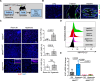
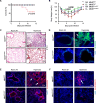
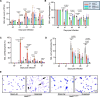
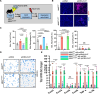
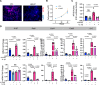
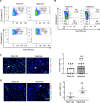
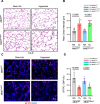
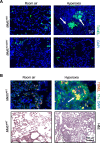
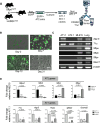
Neutrophils defend against respiratory infections but cause acute lung injury (ALI) when excessively recruited to the lung. Early life environmental factors can shape lung development, but how they impact neutrophil recruitment is not known. We show that exposing newborn mice to hyperoxia increases the number of adult alveolar type 1 (AT1) epithelial cells expressing the proliferation marker Ki-67. Although these cells were not proliferating, they expressed high levels of chemokines that stimulated neutrophil recruitment and ALI when mice were infected with influenza A virus or exposed to lipopolysaccharide (LPS). Neutrophil recruitment and chemokine production were attenuated in Ki-67 hypomorph mice infected with virus or exposed to LPS and enhanced by genetically overexpressing Ki-67 in their lungs. Silencing Ki-67 in a mouse AT1-like cell line reduced basal and IL-1β stimulation of RelA/p65 and NF-κB-dependent transcription of the chemokines Cxcl1 and Cxcl5. Our findings reveal a novel role for Ki-67 to modulate the intensity of epithelial pro-inflammatory signaling, controlling neutrophil recruitment. The severity of respiratory infections may be influenced by mitogens and environmental factors that increase the expression of Ki-67.
Keywords: Acute Lung Injury; Alveolar Epithelial Cells; Influenza A Virus; Mice; Susceptibility.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures





References
-
- Amulic B, Knackstedt SL, Abu Abed U, Deigendesch N, Harbort CJ, Caffrey BE, Brinkmann V, Heppner FL, Hinds PW, Zychlinsky A (2017) Cell-cycle proteins control production of neutrophil extracellular traps. Dev Cell 43:449–462.e445 - PubMed
-
- Andres-Sanchez N, Fisher D, Krasinska L (2022) Physiological functions and roles in cancer of the proliferation marker Ki-67. J Cell Sci 135:jcs258932 - PubMed
MeSH terms
Substances
Grants and funding
- U01 HL148861/HL/NHLBI NIH HHS/United States
- PLBIO18-094/Institut National Du Cancer (INCa)
- R01 ES030300/ES/NIEHS NIH HHS/United States
- R01 HL148825/HL/NHLBI NIH HHS/United States
- ARCPGA12021010002850/Foundation ARC against Cancer
- P30 ES001247/ES/NIEHS NIH HHS/United States
- U01 HL122700/HL/NHLBI NIH HHS/United States
- R01 HL091968/HL/NHLBI NIH HHS/United States
- R01 HL148695/HL/NHLBI NIH HHS/United States
- R01HL148695/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- P30ES001247/HHS | NIH | National Institute of Environmental Health Sciences (DEHS)
- R01ES030300/HHS | NIH | National Institute of Environmental Health Sciences (DEHS)
- R01HL091968,U01HL122700,U01HL148861,R01HL148825/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
LinkOut - more resources
Full Text Sources
Research Materials

