Methodology and application of multiplex PCR-dipstick DNA chromatography for the detection of eight respiratory bacterial pathogens
- PMID: 40496013
- PMCID: PMC12149219
- DOI: 10.3389/fcimb.2025.1558612
Methodology and application of multiplex PCR-dipstick DNA chromatography for the detection of eight respiratory bacterial pathogens
Abstract
Background: Community-acquired pneumonia is primarily caused by Acinetobacter baumannii, Pseudomonas aeruginosa, Klebsiella pneumoniae, Streptococcus pneumoniae, Haemophilus influenzae, Staphylococcus aureus, Mycoplasma pneumoniae, and Chlamydia pneumoniae, leading to severe illness and death in developing countries.
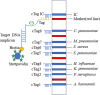
Methods: A rapid, straightforward, sensitive, high-throughput, and precise multiplex PCR-dipstick DNA chromatography assay was devised. This innovative technique was specifically engineered for the immediate and efficient detection of the aforementioned eight respiratory pathogens, with particular emphasis on scenarios involving co-infections. Custom-designed specific primers were employed, wherein the 5' end of the forward primers was tagged with oligonucleotide tags (Tag) and the 5' end of the reverse primers was conjugated with biotin. A C3 spacer was incorporated to bridge the Tag and the forward primer. Complementary oligonucleotides (cTag) corresponding to each of the eight pathogens were immobilized within the test area of the test strip. Meanwhile, biotin was strategically utilized to create an internal control line at the distal end of the test strip. The biotin moiety at the 5' end of the reverse primer was engineered to interact with blue latex microspheres coated with streptavidin, thereby triggering a detectable signal. Following the PCR amplification of the target DNA fragments, during the membrane strip chromatography hybridization process, the Tag- and biotin-labeled target DNA engaged in a dual interaction. First, it bound to the blue latex microspheres via streptavidin-biotin binding, and second, it hybridized with the cTag on the membrane strip. This led to the accumulation of captured blue latex microspheres at both the test line and the internal control line, manifesting as visible blue bands. A total of 186 respiratory sputum or bronchoalveolar lavage fluid specimens were collected and analyzed. The multiplex PCR-dipstick DNA chromatography assay was deployed for detection, while traditional bacterial culture was also carried out in parallel for comparative purposes. To rigorously validate the accuracy of the multiplex PCR-dipstick DNA chromatography assay in identifying PCR products, DNA sequencing was performed on all PCR products derived from the clinical samples.
Results: The multiplex PCR-dipstick DNA chromatography assay demonstrated remarkable efficacy, being capable of specifically discriminating among the eight pathogens within a remarkably short timeframe of 40 minutes. The detection limit for individual bacterial species ranged from 10 to 102 CFU/mL. Notably, no cross-reactions were observed among the eight target bacteria, nor with other representative respiratory bacteria, including Acinetobacter junii, Enterobacter cloacae, Klebsiella oxytoca, Haemophilus parainfluenzae, Pseudomonas fluorescens, Aeromonas hydrophila, and Staphylococcus epidermidis. The concordance between the results obtained from the multiplex PCR-dipstick DNA chromatography assay and those from DNA sequencing was absolute, with a kappa value of 1.00.
Conclusion: A successful multiplex PCR-dipstick DNA chromatography assay was established for the simultaneous detection of eight respiratory bacterial pathogens and was effectively applied in clinical sample analysis. This indicates that this single-use device has promising potential for analyzing the microbial composition related to respiratory infections and is also suitable for small laboratories and field diagnostics.
Keywords: eight respiratory bacterial pathogens; lower respiratory tract infection; multiple detection; multiplex PCR-dipstick DNA chromatography assay; rapid detection.
Copyright © 2025 Hu, Wang and Li.
Conflict of interest statement
Author QL was employed by Guangzhou Baochuang Biotechnology Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Simultaneous Detection of Nine Key Bacterial Respiratory Pathogens Using Luminex xTAG® Technology.Int J Environ Res Public Health. 2017 Feb 23;14(3):223. doi: 10.3390/ijerph14030223. Int J Environ Res Public Health. 2017. PMID: 28241513 Free PMC article.
-
Detection of respiratory bacterial pathogens causing atypical pneumonia by multiplex Lightmix® RT-PCR.Int J Med Microbiol. 2018 Apr;308(3):317-323. doi: 10.1016/j.ijmm.2018.01.010. Epub 2018 Jan 31. Int J Med Microbiol. 2018. PMID: 29397298
-
A LAMP-based system for rapid detection of eight common pathogens causing lower respiratory tract infections.J Microbiol Methods. 2021 Nov;190:106339. doi: 10.1016/j.mimet.2021.106339. Epub 2021 Sep 27. J Microbiol Methods. 2021. PMID: 34592373
-
Design and Evaluation of Multiplex One-Step Reverse Transcription PCR-Dipstick Chromatography Method for the Analysis of Seven Respiratory Pathogens.Curr Microbiol. 2021 Oct;78(10):3656-3666. doi: 10.1007/s00284-021-02621-7. Epub 2021 Aug 2. Curr Microbiol. 2021. PMID: 34338833 Free PMC article.
-
Simultaneous Detection of 13 Key Bacterial Respiratory Pathogens by Combination of Multiplex PCR and Capillary Electrophoresis.Biomed Environ Sci. 2017 Aug;30(8):549-561. doi: 10.3967/bes2017.074. Biomed Environ Sci. 2017. PMID: 28807095
References
-
- Bahavarnia F., Mobed A., Hasanzadeh M., Saadati A., Hassanpour S., Mokhtarzadeh A. (2020). Bio-assay of Acintobacter baumannii using DNA conjugated with gold nano-star: A new platform for microorganism analysis. Enzyme Microb. Technol. 133, 109466. doi: 10.1016/j.enzmictec.2019.109466 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

