Proinflammatory macrophages release CXCL5 to regulate T cell function and limit effects of αPD-1 in steatosis-driven liver cancer
- PMID: 40496444
- PMCID: PMC12151196
- DOI: 10.1016/j.jhepr.2025.101385
Proinflammatory macrophages release CXCL5 to regulate T cell function and limit effects of αPD-1 in steatosis-driven liver cancer
Abstract
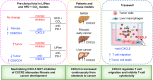
Background & aims: Steatosis is a comorbid factor for cancer development. Patients with steatosis do not respond well to current immune checkpoint therapy (CPI) treatment. We explored the roles of neutrophil-activating chemokines (NACs) in the response of steatosis/liver cancer to CPI.
Methods: We used a steatosis-driven liver cancer model induced by the deletion of Pten in the liver (LiPten) and a high-fat diet + carbon tetrachloride (CCl4) fibrosis model to study the effects of targeting CXCL5. We also studied the role of CXCL5 in the liver immune microenvironment in vitro and in vivo. ANOVA/t tests were used for data analysis.
Results: Using LiPten steatosis-tumor mice, we identified CXCL5 as the NAC most robustly upregulated as steatosis progresses to cancer (>100 fold, n = 6-11). We also validated this observation in patient samples. When used together with αPD-1, inhibiting the NAC receptor CXCR2 promoted (100% vs. 80% in untreated LiPten mice), whereas anti-CXCL5 suppressed (25%), tumor progression (n = 4-6) suggesting unique functions of CXCL5 independent of CXCR2. Similar effects were observed for anti-CXCL5 (0/4 with fibrosis) vs. CXCR2 inhibition (4/4 with fibrosis) of fibrosis in the HFD + CCl4 model. Using a Transwell assay, we identified a novel inhibitory function of CXCL5 in the recruitment of CD4+ T cells (p <0.02, n = 4) and potentiation of CD8+ T cell cytotoxicity (p <0.001, n = 4). In vivo, we showed that neutralizing CXCL5 increased the CD8/CD4 ratio (p = 0.03 and 0.07) and synergized with αPD-1 for its anti-tumor and anti-fibrosis activity (n = 4-6).
Conclusions: Our discovery of the novel inhibitory role of CXCL5 in T cells suggests that NACs have additional functions in modulating the immune system beyond neutrophil chemotaxis. The discovery of this novel CXCL5 role presents additional therapeutical targets alongside current immune checkpoint therapy.
Impact and implications: In this study, we investigated the role of CXCL5 in the progression from steatosis to liver cancer. We uncovered a novel inhibitory role of CXCL5 in T cell recruitment, with implications for NAC-targeted therapy and immune checkpoint synergy in liver cancer. We believe our findings will be of interest to physicians, researchers, and patients interested in therapeutic development and translational research in liver disease.
Keywords: Fibrosis; HCC; NAFLD/NASH; PTEN.
© 2025 The Authors.
Conflict of interest statement
The authors declare no conflicts of interest that pertain to this work.
Figures








References
-
- Ramachandra N., Gupta M., Schwartz L., et al. Role of IL8 in myeloid malignancies. Leuk Lymphoma. 2023;64:1742–1751. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

