Updated ACMG/AMP specifications for variant interpretation and gene curations from the ClinGen RASopathy expert panels
- PMID: 40496714
- PMCID: PMC12151217
- DOI: 10.1016/j.gimo.2025.103430
Updated ACMG/AMP specifications for variant interpretation and gene curations from the ClinGen RASopathy expert panels
Abstract
Purpose: The ClinGen RASopathy (RAS) Variant Curation Expert Panel (VCEP) previously established RASopathy specifications to the American College of Medical Genetics and Genomics (ACMG) and Association of Molecular Pathology (AMP) variant classification framework for more consistent and accurate variant classification. Advances in the understanding of RASopathies and new clinical genetic testing algorithms required updated specifications.
Methods: The RAS Gene Curation Expert Panel recurated 6 gene-disease relationships, and the RAS VCEP evaluated the previous specifications to develop updated RASopathy specifications for the ACMG/AMP framework. The performance of these updated specifications was tested by reassessing 59 previously classified variants and 88 new pilot variants.
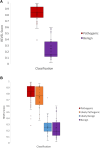
Results: Five gene-disease relationships were upgraded to Definitive, whereas 1 was upgraded to Moderate. Updated specifications were applied to 11 ACMG/AMP criteria for disorders with a dominant inheritance, 3 criteria for recessive inheritance, and 4 criteria to align with recommendations from the ClinGen Sequence Variant Interpretation Working Group. Assessment of variants demonstrated no major shifts in classifications compared with previous RAS VCEP or ClinVar classifications.
Conclusion: Updated RASopathy specifications improve the classification of variants associated with recessive disease and observed in exome/genome cases. Most of these specifications may also be used as a baseline for other rare Mendelian disorders.
Keywords: ClinGen; Noonan; RASopathy; Ras/MAPK; Variant interpretation.
© 2025 The Authors.
Conflict of interest statement
Bradley Williams and Lisa M. Vincent work for a for-profit genetic testing center. Bruce D. Gelb receives royalties from GeneDx, Correlegan, LabCorp, and Prevention Genetics; consulted for Day One BioPharmaceuticals, BioMarin; and had sponsored research agreements from Day One BioPharmaceuticals, Onconova. All other authors declare no conflicts of interest.
Figures
References
-
- Seifert B.A., McGlaughon J.L., Jackson S.A., et al. Determining the clinical validity of hereditary colorectal cancer and polyposis susceptibility genes using the Clinical Genome Resource Clinical Validity Framework. Genet Med. 2019;21(7):1507–1516. doi: 10.1038/s41436-018-0373-1. - DOI - PMC - PubMed
Grants and funding
- U24 CA258115/CA/NCI NIH HHS/United States
- U24 HG006834/HG/NHGRI NIH HHS/United States
- U24 HD104590/HD/NICHD NIH HHS/United States
- U24 CA258119/CA/NCI NIH HHS/United States
- U24 HD093483/HD/NICHD NIH HHS/United States
- U41 HG009649/HG/NHGRI NIH HHS/United States
- U24 EY033699/EY/NEI NIH HHS/United States
- U24 HD104588/HD/NICHD NIH HHS/United States
- U41 HG009650/HG/NHGRI NIH HHS/United States
- U24 HD108087/HD/NICHD NIH HHS/United States
- U24 EY032451/EY/NEI NIH HHS/United States
- U24 CA258118/CA/NCI NIH HHS/United States
- U41 HG006834/HG/NHGRI NIH HHS/United States
- U24 NS120858/NS/NINDS NIH HHS/United States
- U24 HD104592/HD/NICHD NIH HHS/United States
- U24 HD104591/HD/NICHD NIH HHS/United States
- U24 NS120854/NS/NINDS NIH HHS/United States
- U24 CA258058/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources

