Clinical evaluation of two pathogen enrichment approaches for next-generation sequencing in the diagnosis of lower respiratory tract infections
- PMID: 40497682
- PMCID: PMC12210998
- DOI: 10.1128/spectrum.00922-25
Clinical evaluation of two pathogen enrichment approaches for next-generation sequencing in the diagnosis of lower respiratory tract infections
Abstract
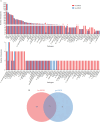
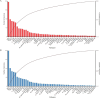
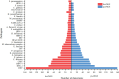
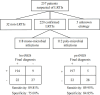
The underdevelopment of microbiological tests has contributed to diagnostic delay and inappropriate use of antibiotics in patients with lower respiratory tract infections, which is ranked as the seventh leading cause of death globally. Next-generation sequencing (NGS) has emerged as a promising platform for the diagnosis of infectious diseases, albeit with high costs and challenges in result interpretation. Here we evaluated two NGS-based pathogen detection assays for the etiological diagnosis of pneumonia in a prospective cohort of 257 patients. Both assays utilized multiplex polymerase chain reaction (PCR) for pathogen enrichment. One assay was designed to promiscuously amplify and identify more than 1,000 pathogens (broad-spectrum targeted next-generation sequencing [bs-tNGS]), while the other specifically targeted 194 pathogens (pathogen-specific targeted next-generation sequencing [ps-tNGS]). The analytical and diagnostic performances of both assays were compared using a composite clinical reference standard. The specificity of ps-tNGS was higher than that of bs-tNGS (84.85% vs. 75.00%), while the sensitivities of both assays were similar (>89%). In addition, a significant overlap in the frequently detected pathogens by the two methods was observed. Moreover, the enrichment of pathogens via multiplex PCR for ps-tNGS has alleviated the requirement for deep sequencing in the shotgun metagenomic workflows and thus dramatically lowered the assay cost. This study demonstrated that ps-tNGS achieved a better overall diagnostic performance and may potentially replace bs-tNGS in the clinical application.IMPORTANCEMicrobial enrichment in metagenomic next-generation sequencing has been achieved through differential cell lysis, but the results varied, depending on experimental procedures and sample types. Therefore, direct enrichment of pathogen DNA/RNA was attempted via multiplex PCR or hybrid probe capture (targeted next-generation sequencing [tNGS]). We evaluated two enrichment methods based on multiplex PCR. One method utilized a primer design strategy to amplify over 1,000 respiratory pathogens (bs-tNGS), while the other specifically targeted 194 pathogens (ps-tNGS). Our findings disavowed the notion that "the more, the better" in tNGS workflows, since ps-tNGS exhibited equivalent sensitivity and, notably, higher specificity than bs-tNGS in a prospective cohort of 257 patients who were suspected of having pneumonia. In future evaluations of tNGS assays, researchers should pay more attention to diagnostic specificity, rather than focusing solely on sensitivity, since a low specificity may potentially lead to misdiagnosis and overuse of antibiotics in cases of non-infectious diseases.
Keywords: etiological diagnosis; lower respiratory tract infection; multiplex PCR; pathogen enrichment; targeted next-generation sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- GBD 2021 Causes of Death Collaborators . 2024. Global burden of 288 causes of death and life expectancy decomposition in 204 countries and territories and 811 subnational locations, 1990-2021: a systematic analysis for the Global Burden of Disease Study 2021. Lancet 403:2100–2132. doi: 10.1016/S0140-6736(24)00367-2 - DOI - PMC - PubMed
-
- Holter JC, Müller F, Bjørang O, Samdal HH, Marthinsen JB, Jenum PA, Ueland T, Frøland SS, Aukrust P, Husebye E, Heggelund L. 2015. Etiology of community-acquired pneumonia and diagnostic yields of microbiological methods: a 3-year prospective study in Norway. BMC Infect Dis 15:64. doi: 10.1186/s12879-015-0803-5 - DOI - PMC - PubMed
-
- Vos LM, Bruyndonckx R, Zuithoff NPA, Little P, Oosterheert JJ, Broekhuizen BDL, Lammens C, Loens K, Viveen M, Butler CC, Crook D, Zlateva K, Goossens H, Claas ECJ, Ieven M, Van Loon AM, Verheij TJM, Coenjaerts FEJ, GRACE Consortium . 2021. Lower respiratory tract infection in the community: associations between viral aetiology and illness course. Clin Microbiol Infect 27:96–104. doi: 10.1016/j.cmi.2020.03.023 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

