Transcriptomic Analyses of Ovarian Clear Cell Carcinoma Spheroids Reveal Distinct Proliferative Phenotypes and Therapeutic Vulnerabilities
- PMID: 40497961
- PMCID: PMC12154277
- DOI: 10.3390/cells14110785
Transcriptomic Analyses of Ovarian Clear Cell Carcinoma Spheroids Reveal Distinct Proliferative Phenotypes and Therapeutic Vulnerabilities
Abstract
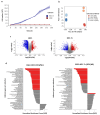
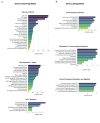
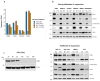
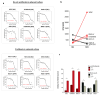
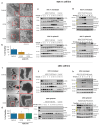
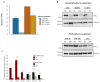
Cancer cell spheroids autonomously form in the ascites fluid and are considered a conduit for epithelial ovarian cancer metastasis within the peritoneal cavity. Spheroids are homotypic, avascular 3D structures that acquire resistance to anoikis to remain viable after cellular detachment. We used in vitro spheroid model systems to interrogate pathways critical for spheroid cell proliferation, distinct from those driving monolayer cancer cell proliferation. Using the 105C and KOC-7c human ovarian clear cell carcinoma (OCCC) cell lines, which have distinct proliferative phenotypes as spheroids but the same prototypical OCCC gene mutation profile of constitutively activated AKT signaling with the loss of ARID1A, we revealed therapeutic targets that efficiently kill cells in spheroids. RNA-seq analyses compared the transcriptome of 3-day monolayer and spheroid cells from these lines and identified the characteristics of dormant spheroid cell survival, which included the G2/M checkpoint, autophagy, and other stress pathways induced in 105C spheroids, in sharp contrast to the proliferating spheroid cells of the KOC-7c cell line. Next, we assessed levels of various G2/M checkpoint regulators and found a consistent reduction in steady-state levels of checkpoint regulators in dormant spheroid cells, but not proliferative spheroids. Our studies showed that proliferative spheroid cells were sensitive to Wee1 inhibition by AZD1775, but the dormant spheroid cells showed a degree of resistance to AZD1775, both in terms of EC50 values and spheroid reattachment abilities. Thus, we identified biomarkers of dormant spheroids, including the G2/M checkpoint regulators Wee1, Cdc25c, and PLK1, and showed that, when compared to proliferating spheroid cells, the transcriptome of dormant OCCC spheroids is a source of therapeutic targets.
Keywords: AZD1775; G2/M; cancer; cell line; clear cell; dormancy; ovarian; spheroid.
Conflict of interest statement
The authors declare no conflicts of interest for this study.
Figures

References
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

